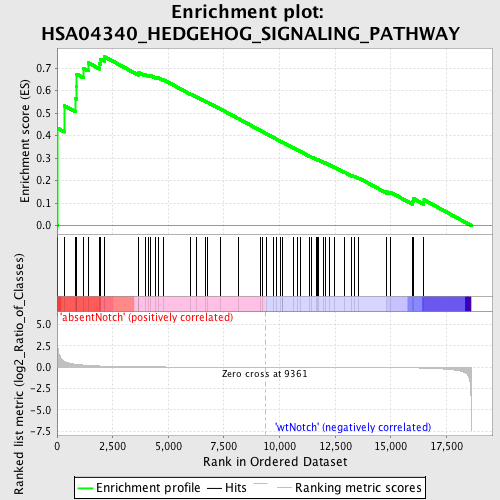
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch_repos |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#absentNotch_versus_wtNotch_repos |
| Upregulated in class | absentNotch |
| GeneSet | HSA04340_HEDGEHOG_SIGNALING_PATHWAY |
| Enrichment Score (ES) | 0.75038004 |
| Normalized Enrichment Score (NES) | 1.586541 |
| Nominal p-value | 0.0018348624 |
| FDR q-value | 0.33313265 |
| FWER p-Value | 0.984 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CSNK1G2 | 3060095 4730037 | 30 | 2.476 | 0.4326 | Yes | ||
| 2 | WNT8B | 4560301 | 336 | 0.663 | 0.5325 | Yes | ||
| 3 | CSNK1D | 4280280 4480167 6450600 | 822 | 0.335 | 0.5650 | Yes | ||
| 4 | PRKACB | 4210170 | 882 | 0.318 | 0.6176 | Yes | ||
| 5 | CSNK1E | 2850347 5050093 6110301 | 887 | 0.316 | 0.6729 | Yes | ||
| 6 | WNT6 | 4570364 | 1181 | 0.244 | 0.6999 | Yes | ||
| 7 | GSK3B | 5360348 | 1401 | 0.203 | 0.7237 | Yes | ||
| 8 | WNT5B | 5080162 | 1912 | 0.135 | 0.7199 | Yes | ||
| 9 | PRKACA | 2640731 4050048 | 1953 | 0.129 | 0.7404 | Yes | ||
| 10 | BMP7 | 870039 | 2141 | 0.114 | 0.7504 | Yes | ||
| 11 | SMO | 5340538 6350132 | 3643 | 0.038 | 0.6762 | No | ||
| 12 | RAB23 | 2970075 | 3674 | 0.038 | 0.6812 | No | ||
| 13 | WNT2B | 3130088 5960575 | 3960 | 0.031 | 0.6713 | No | ||
| 14 | GLI2 | 3060632 | 4114 | 0.029 | 0.6681 | No | ||
| 15 | BMP8A | 1990537 | 4206 | 0.027 | 0.6680 | No | ||
| 16 | WNT9A | 3830711 | 4432 | 0.024 | 0.6601 | No | ||
| 17 | CSNK1A1 | 2340427 | 4566 | 0.022 | 0.6568 | No | ||
| 18 | WNT3A | 6350348 | 4801 | 0.020 | 0.6476 | No | ||
| 19 | LRP2 | 1400204 3290706 | 6016 | 0.011 | 0.5842 | No | ||
| 20 | PTCH2 | 2900092 | 6274 | 0.010 | 0.5720 | No | ||
| 21 | SHH | 5570400 | 6670 | 0.008 | 0.5522 | No | ||
| 22 | WNT10A | 2100706 | 6754 | 0.008 | 0.5490 | No | ||
| 23 | WNT4 | 4150619 | 7335 | 0.006 | 0.5188 | No | ||
| 24 | HHIP | 6940438 | 8160 | 0.003 | 0.4750 | No | ||
| 25 | BMP5 | 4670048 | 9144 | 0.001 | 0.4221 | No | ||
| 26 | WNT3 | 2360685 | 9219 | 0.000 | 0.4182 | No | ||
| 27 | WNT7B | 3520039 | 9400 | -0.000 | 0.4085 | No | ||
| 28 | BMP6 | 2100128 | 9709 | -0.001 | 0.3921 | No | ||
| 29 | WNT8A | 4150014 5390731 | 9849 | -0.001 | 0.3848 | No | ||
| 30 | GLI1 | 4560176 | 10058 | -0.002 | 0.3739 | No | ||
| 31 | DHH | 2120433 | 10135 | -0.002 | 0.3702 | No | ||
| 32 | WNT9B | 50095 | 10145 | -0.002 | 0.3700 | No | ||
| 33 | FBXW11 | 6450632 | 10645 | -0.003 | 0.3438 | No | ||
| 34 | ZIC2 | 1410408 | 10810 | -0.004 | 0.3356 | No | ||
| 35 | IHH | 1980735 | 10950 | -0.004 | 0.3289 | No | ||
| 36 | PRKX | 2480390 2810026 | 11339 | -0.006 | 0.3090 | No | ||
| 37 | WNT7A | 1170315 | 11450 | -0.006 | 0.3041 | No | ||
| 38 | WNT2 | 4610020 | 11667 | -0.007 | 0.2937 | No | ||
| 39 | BMP8B | 3290102 | 11725 | -0.007 | 0.2918 | No | ||
| 40 | PTCH1 | 430278 | 11736 | -0.007 | 0.2925 | No | ||
| 41 | GAS1 | 2120504 | 11959 | -0.008 | 0.2820 | No | ||
| 42 | BMP2 | 6620687 | 12048 | -0.008 | 0.2787 | No | ||
| 43 | GLI3 | 5690148 | 12233 | -0.009 | 0.2703 | No | ||
| 44 | WNT16 | 3870563 4200687 | 12456 | -0.010 | 0.2602 | No | ||
| 45 | BMP4 | 380113 | 12935 | -0.013 | 0.2367 | No | ||
| 46 | WNT11 | 1230278 | 13243 | -0.015 | 0.2227 | No | ||
| 47 | CSNK1G1 | 840082 1230575 3940647 | 13361 | -0.016 | 0.2192 | No | ||
| 48 | WNT1 | 4780148 | 13567 | -0.018 | 0.2113 | No | ||
| 49 | BTRC | 3170131 | 14812 | -0.035 | 0.1505 | No | ||
| 50 | WNT5A | 840685 3120152 | 14967 | -0.039 | 0.1491 | No | ||
| 51 | SUFU | 6650181 | 15972 | -0.077 | 0.1086 | No | ||
| 52 | WNT10B | 510050 | 16005 | -0.079 | 0.1208 | No | ||
| 53 | CSNK1G3 | 110450 | 16490 | -0.113 | 0.1145 | No |