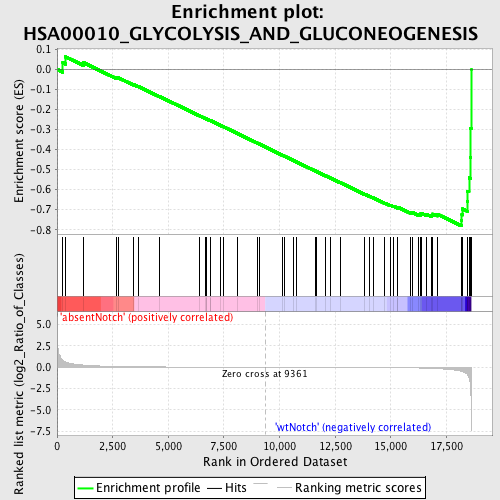
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch_repos |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#absentNotch_versus_wtNotch_repos |
| Upregulated in class | wtNotch |
| GeneSet | HSA00010_GLYCOLYSIS_AND_GLUCONEOGENESIS |
| Enrichment Score (ES) | -0.7812155 |
| Normalized Enrichment Score (NES) | -1.7122606 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.09135553 |
| FWER p-Value | 0.505 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ALDH3B1 | 4210010 6940403 | 257 | 0.790 | 0.0328 | No | ||
| 2 | HK2 | 2640722 | 381 | 0.613 | 0.0624 | No | ||
| 3 | ADH1A | 3170427 | 1189 | 0.243 | 0.0332 | No | ||
| 4 | G6PC | 430093 | 2666 | 0.077 | -0.0417 | No | ||
| 5 | ACSS2 | 1780435 3170224 | 2754 | 0.073 | -0.0421 | No | ||
| 6 | ALDOC | 450121 610427 | 3427 | 0.044 | -0.0757 | No | ||
| 7 | ALDH2 | 4230019 | 3635 | 0.038 | -0.0846 | No | ||
| 8 | GALM | 730368 | 4581 | 0.022 | -0.1342 | No | ||
| 9 | PKLR | 1170400 2470114 | 6390 | 0.009 | -0.2310 | No | ||
| 10 | ALDH7A1 | 4050446 | 6685 | 0.008 | -0.2464 | No | ||
| 11 | ACYP2 | 2510605 | 6711 | 0.008 | -0.2473 | No | ||
| 12 | PFKL | 6200167 | 6723 | 0.008 | -0.2474 | No | ||
| 13 | ALDH9A1 | 4810047 | 6881 | 0.007 | -0.2555 | No | ||
| 14 | ALDH1A3 | 2100270 | 6900 | 0.007 | -0.2560 | No | ||
| 15 | PDHA2 | 2630438 | 7331 | 0.006 | -0.2788 | No | ||
| 16 | LDHAL6B | 5130280 | 7470 | 0.005 | -0.2860 | No | ||
| 17 | ALDOB | 4730324 | 8087 | 0.003 | -0.3189 | No | ||
| 18 | PDHB | 70215 610086 | 9006 | 0.001 | -0.3683 | No | ||
| 19 | PGM3 | 2570465 | 9113 | 0.001 | -0.3740 | No | ||
| 20 | GCK | 2370273 | 10132 | -0.002 | -0.4287 | No | ||
| 21 | GAPDHS | 2690463 | 10198 | -0.002 | -0.4321 | No | ||
| 22 | ACYP1 | 6380403 | 10629 | -0.003 | -0.4551 | No | ||
| 23 | PGK2 | 2650091 | 10781 | -0.004 | -0.4630 | No | ||
| 24 | BPGM | 5080520 | 11595 | -0.007 | -0.5064 | No | ||
| 25 | PGM1 | 3840408 | 11642 | -0.007 | -0.5085 | No | ||
| 26 | G6PC2 | 1660114 5220070 | 12082 | -0.008 | -0.5316 | No | ||
| 27 | ADH4 | 6840301 | 12290 | -0.009 | -0.5422 | No | ||
| 28 | ADHFE1 | 1990673 6840022 | 12743 | -0.012 | -0.5659 | No | ||
| 29 | FBP2 | 1580193 | 13817 | -0.020 | -0.6225 | No | ||
| 30 | ADH7 | 60465 | 14051 | -0.023 | -0.6337 | No | ||
| 31 | ENO2 | 2320068 | 14205 | -0.025 | -0.6405 | No | ||
| 32 | DLAT | 430452 | 14734 | -0.034 | -0.6669 | No | ||
| 33 | ENO1 | 5340128 | 14984 | -0.040 | -0.6780 | No | ||
| 34 | HK1 | 4280402 | 15118 | -0.043 | -0.6826 | No | ||
| 35 | PDHA1 | 5550397 | 15310 | -0.050 | -0.6899 | No | ||
| 36 | LDHC | 2030458 6100463 | 15315 | -0.050 | -0.6872 | No | ||
| 37 | PGAM2 | 3610605 | 15878 | -0.073 | -0.7132 | No | ||
| 38 | GAPDH | 110022 430039 6220161 | 15980 | -0.078 | -0.7140 | No | ||
| 39 | DLD | 4150403 6590341 | 16231 | -0.092 | -0.7220 | No | ||
| 40 | HK3 | 3190288 6510142 | 16325 | -0.099 | -0.7212 | No | ||
| 41 | ALDH3A1 | 580095 | 16392 | -0.104 | -0.7186 | No | ||
| 42 | ACSS1 | 6580037 | 16601 | -0.119 | -0.7227 | No | ||
| 43 | PFKM | 1990156 5720168 | 16826 | -0.138 | -0.7266 | No | ||
| 44 | LDHA | 2190594 | 16890 | -0.145 | -0.7215 | No | ||
| 45 | PGAM1 | 2570133 | 17077 | -0.165 | -0.7217 | No | ||
| 46 | ADH5 | 6290100 6380711 | 18182 | -0.481 | -0.7528 | Yes | ||
| 47 | ALDOA | 6290672 | 18195 | -0.486 | -0.7248 | Yes | ||
| 48 | PKM2 | 6520403 70500 | 18229 | -0.514 | -0.6962 | Yes | ||
| 49 | FBP1 | 1470762 | 18434 | -0.818 | -0.6588 | Yes | ||
| 50 | ENO3 | 5270136 | 18453 | -0.871 | -0.6083 | Yes | ||
| 51 | TPI1 | 1500215 2100154 | 18514 | -1.197 | -0.5408 | Yes | ||
| 52 | PFKP | 70138 6760040 1170278 | 18572 | -1.792 | -0.4381 | Yes | ||
| 53 | ALDH1B1 | 610195 | 18591 | -2.472 | -0.2930 | Yes | ||
| 54 | PGK1 | 1570494 630300 | 18612 | -4.983 | 0.0002 | Yes |

