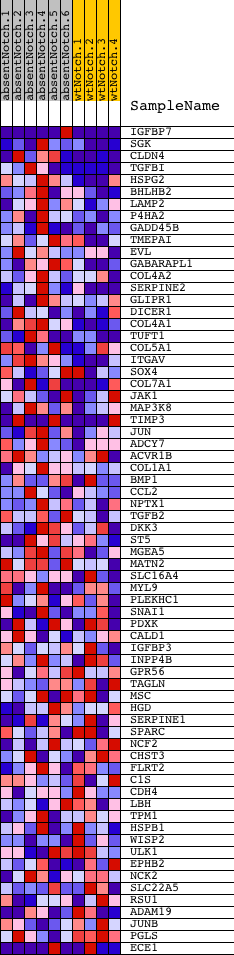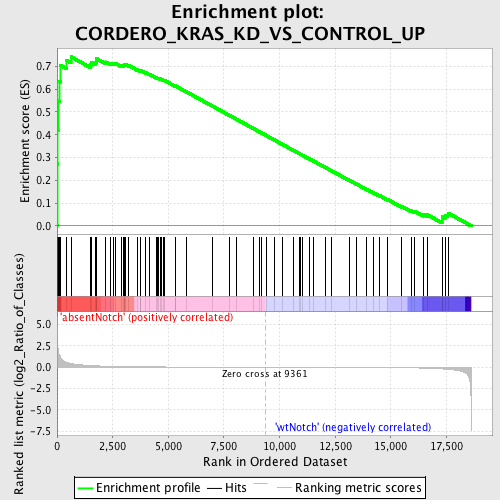
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch.phenotype_absentNotch_versus_wtNotch.cls #absentNotch_versus_wtNotch_repos |
| Phenotype | phenotype_absentNotch_versus_wtNotch.cls#absentNotch_versus_wtNotch_repos |
| Upregulated in class | absentNotch |
| GeneSet | CORDERO_KRAS_KD_VS_CONTROL_UP |
| Enrichment Score (ES) | 0.7419832 |
| Normalized Enrichment Score (NES) | 1.674564 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.13120864 |
| FWER p-Value | 0.425 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | IGFBP7 | 520411 3060110 5290152 | 6 | 4.192 | 0.2723 | Yes | ||
| 2 | SGK | 1400131 2480056 | 32 | 2.307 | 0.4210 | Yes | ||
| 3 | CLDN4 | 4920739 | 45 | 1.980 | 0.5491 | Yes | ||
| 4 | TGFBI | 2060446 6900112 | 113 | 1.357 | 0.6338 | Yes | ||
| 5 | HSPG2 | 2510687 6220750 | 158 | 1.113 | 0.7038 | Yes | ||
| 6 | BHLHB2 | 7040603 | 437 | 0.560 | 0.7252 | Yes | ||
| 7 | LAMP2 | 1230402 1980373 | 626 | 0.413 | 0.7420 | Yes | ||
| 8 | P4HA2 | 1090546 | 1497 | 0.187 | 0.7073 | No | ||
| 9 | GADD45B | 2350408 | 1528 | 0.181 | 0.7175 | No | ||
| 10 | TMEPAI | 1340253 | 1734 | 0.154 | 0.7164 | No | ||
| 11 | EVL | 1740113 | 1765 | 0.151 | 0.7246 | No | ||
| 12 | GABARAPL1 | 2810458 | 1774 | 0.150 | 0.7340 | No | ||
| 13 | COL4A2 | 2350619 | 2179 | 0.111 | 0.7194 | No | ||
| 14 | SERPINE2 | 2810390 | 2384 | 0.095 | 0.7146 | No | ||
| 15 | GLIPR1 | 3360750 | 2537 | 0.085 | 0.7120 | No | ||
| 16 | DICER1 | 540286 | 2636 | 0.079 | 0.7118 | No | ||
| 17 | COL4A1 | 1740575 | 2884 | 0.066 | 0.7028 | No | ||
| 18 | TUFT1 | 540047 2370010 | 2981 | 0.062 | 0.7016 | No | ||
| 19 | COL5A1 | 2230050 | 3006 | 0.060 | 0.7043 | No | ||
| 20 | ITGAV | 2340020 | 3029 | 0.059 | 0.7069 | No | ||
| 21 | SOX4 | 2260091 | 3095 | 0.056 | 0.7071 | No | ||
| 22 | COL7A1 | 1230168 2190072 | 3204 | 0.052 | 0.7046 | No | ||
| 23 | JAK1 | 5910746 | 3624 | 0.039 | 0.6846 | No | ||
| 24 | MAP3K8 | 2940286 | 3748 | 0.036 | 0.6803 | No | ||
| 25 | TIMP3 | 1450504 1980270 | 3764 | 0.036 | 0.6818 | No | ||
| 26 | JUN | 840170 | 3990 | 0.031 | 0.6717 | No | ||
| 27 | ADCY7 | 6290520 7560739 | 4160 | 0.028 | 0.6644 | No | ||
| 28 | ACVR1B | 3610446 5570195 | 4444 | 0.024 | 0.6507 | No | ||
| 29 | COL1A1 | 730020 | 4506 | 0.023 | 0.6489 | No | ||
| 30 | BMP1 | 380594 2940576 3710593 | 4556 | 0.022 | 0.6477 | No | ||
| 31 | CCL2 | 4760019 | 4626 | 0.022 | 0.6454 | No | ||
| 32 | NPTX1 | 3710736 | 4705 | 0.021 | 0.6425 | No | ||
| 33 | TGFB2 | 4920292 | 4792 | 0.020 | 0.6392 | No | ||
| 34 | DKK3 | 6450072 7050128 | 4848 | 0.019 | 0.6374 | No | ||
| 35 | ST5 | 3780204 | 5299 | 0.015 | 0.6142 | No | ||
| 36 | MGEA5 | 6100014 | 5305 | 0.015 | 0.6149 | No | ||
| 37 | MATN2 | 460215 | 5829 | 0.012 | 0.5875 | No | ||
| 38 | SLC16A4 | 2450500 | 6996 | 0.007 | 0.5251 | No | ||
| 39 | MYL9 | 4210750 7050138 | 7735 | 0.004 | 0.4856 | No | ||
| 40 | PLEKHC1 | 2650066 | 8081 | 0.003 | 0.4672 | No | ||
| 41 | SNAI1 | 6590253 | 8823 | 0.001 | 0.4273 | No | ||
| 42 | PDXK | 6840066 | 9099 | 0.001 | 0.4125 | No | ||
| 43 | CALD1 | 1770129 1940397 | 9165 | 0.001 | 0.4091 | No | ||
| 44 | IGFBP3 | 2370500 | 9391 | -0.000 | 0.3969 | No | ||
| 45 | INPP4B | 6200504 | 9782 | -0.001 | 0.3760 | No | ||
| 46 | GPR56 | 1190494 | 10125 | -0.002 | 0.3577 | No | ||
| 47 | TAGLN | 6660280 | 10608 | -0.003 | 0.3319 | No | ||
| 48 | MSC | 5080139 | 10890 | -0.004 | 0.3170 | No | ||
| 49 | HGD | 460390 | 10943 | -0.004 | 0.3145 | No | ||
| 50 | SERPINE1 | 4210403 | 11015 | -0.005 | 0.3110 | No | ||
| 51 | SPARC | 1690086 | 11335 | -0.006 | 0.2941 | No | ||
| 52 | NCF2 | 540129 2370441 2650133 | 11503 | -0.006 | 0.2855 | No | ||
| 53 | CHST3 | 2370131 | 12070 | -0.008 | 0.2556 | No | ||
| 54 | FLRT2 | 4200035 | 12343 | -0.010 | 0.2415 | No | ||
| 55 | C1S | 840184 6840114 | 13148 | -0.014 | 0.1991 | No | ||
| 56 | CDH4 | 6550368 | 13443 | -0.017 | 0.1843 | No | ||
| 57 | LBH | 70093 | 13909 | -0.021 | 0.1606 | No | ||
| 58 | TPM1 | 130673 | 14232 | -0.025 | 0.1449 | No | ||
| 59 | HSPB1 | 6760435 | 14489 | -0.029 | 0.1331 | No | ||
| 60 | WISP2 | 5550411 | 14854 | -0.036 | 0.1158 | No | ||
| 61 | ULK1 | 6100315 | 15490 | -0.056 | 0.0852 | No | ||
| 62 | EPHB2 | 1090288 1410377 2100541 | 15929 | -0.075 | 0.0665 | No | ||
| 63 | NCK2 | 2510010 | 16060 | -0.083 | 0.0649 | No | ||
| 64 | SLC22A5 | 2510358 2510451 | 16462 | -0.110 | 0.0504 | No | ||
| 65 | RSU1 | 4850372 | 16630 | -0.121 | 0.0493 | No | ||
| 66 | ADAM19 | 5340075 | 17304 | -0.203 | 0.0262 | No | ||
| 67 | JUNB | 4230048 | 17305 | -0.203 | 0.0394 | No | ||
| 68 | PGLS | 2120324 | 17439 | -0.228 | 0.0471 | No | ||
| 69 | ECE1 | 3870093 3940551 | 17570 | -0.250 | 0.0564 | No |