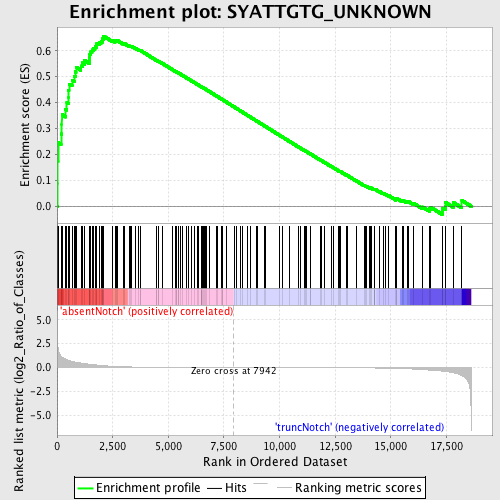
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch.phenotype_absentNotch_versus_truncNotch.cls #absentNotch_versus_truncNotch_repos |
| Phenotype | phenotype_absentNotch_versus_truncNotch.cls#absentNotch_versus_truncNotch_repos |
| Upregulated in class | absentNotch |
| GeneSet | SYATTGTG_UNKNOWN |
| Enrichment Score (ES) | 0.65644467 |
| Normalized Enrichment Score (NES) | 1.6617442 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.015289468 |
| FWER p-Value | 0.133 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | HNRPH1 | 1170086 3140546 3290471 6110184 6110373 | 17 | 2.557 | 0.0876 | Yes | ||
| 2 | CUTL1 | 1570113 1770091 1850273 2100286 5340605 | 19 | 2.519 | 0.1747 | Yes | ||
| 3 | ILF3 | 940722 3190647 6520110 | 40 | 2.079 | 0.2456 | Yes | ||
| 4 | SFRS6 | 60224 | 177 | 1.185 | 0.2792 | Yes | ||
| 5 | ABI1 | 2850152 | 216 | 1.099 | 0.3152 | Yes | ||
| 6 | KCNF1 | 2650687 | 219 | 1.092 | 0.3529 | Yes | ||
| 7 | MAPK6 | 6760520 | 376 | 0.875 | 0.3747 | Yes | ||
| 8 | FOXP1 | 6510433 | 441 | 0.810 | 0.3993 | Yes | ||
| 9 | DDX3X | 2190020 | 518 | 0.751 | 0.4212 | Yes | ||
| 10 | PIK3R3 | 1770333 | 523 | 0.744 | 0.4467 | Yes | ||
| 11 | PDAP1 | 1410735 | 535 | 0.739 | 0.4717 | Yes | ||
| 12 | AHCYL1 | 540609 | 674 | 0.646 | 0.4866 | Yes | ||
| 13 | ELOVL5 | 3800170 | 772 | 0.598 | 0.5020 | Yes | ||
| 14 | KLF13 | 4670563 | 830 | 0.570 | 0.5187 | Yes | ||
| 15 | SOX4 | 2260091 | 850 | 0.562 | 0.5371 | Yes | ||
| 16 | EIF4G1 | 4070446 | 1073 | 0.470 | 0.5414 | Yes | ||
| 17 | TRIM39 | 6550441 | 1140 | 0.447 | 0.5533 | Yes | ||
| 18 | RANBP10 | 2340184 | 1237 | 0.413 | 0.5624 | Yes | ||
| 19 | BCL2L11 | 780044 4200601 | 1444 | 0.349 | 0.5633 | Yes | ||
| 20 | YTHDF2 | 4060332 4610279 | 1451 | 0.348 | 0.5750 | Yes | ||
| 21 | WASF1 | 2060484 | 1472 | 0.342 | 0.5858 | Yes | ||
| 22 | CGGBP1 | 4810053 | 1509 | 0.334 | 0.5954 | Yes | ||
| 23 | BCL7A | 4560095 | 1569 | 0.316 | 0.6031 | Yes | ||
| 24 | BCL2L1 | 1580452 4200152 5420484 | 1636 | 0.296 | 0.6098 | Yes | ||
| 25 | DLX1 | 360168 | 1704 | 0.279 | 0.6158 | Yes | ||
| 26 | MAPK10 | 6110193 | 1760 | 0.267 | 0.6221 | Yes | ||
| 27 | ARIH1 | 6900025 | 1787 | 0.260 | 0.6297 | Yes | ||
| 28 | RPL30 | 4810369 | 1918 | 0.231 | 0.6306 | Yes | ||
| 29 | CLU | 5420075 | 1974 | 0.216 | 0.6351 | Yes | ||
| 30 | PLAGL2 | 1770148 | 2022 | 0.206 | 0.6397 | Yes | ||
| 31 | ORMDL3 | 1340711 | 2040 | 0.201 | 0.6457 | Yes | ||
| 32 | YWHAZ | 1230717 | 2082 | 0.195 | 0.6503 | Yes | ||
| 33 | NSD1 | 4560519 4780672 | 2092 | 0.193 | 0.6564 | Yes | ||
| 34 | POLR3E | 380152 | 2470 | 0.131 | 0.6406 | No | ||
| 35 | MYO1C | 3190398 | 2603 | 0.118 | 0.6375 | No | ||
| 36 | RNGTT | 780152 780746 1090471 | 2649 | 0.113 | 0.6390 | No | ||
| 37 | NDST2 | 6290711 | 2711 | 0.106 | 0.6394 | No | ||
| 38 | GUCA1A | 6020338 | 2998 | 0.080 | 0.6267 | No | ||
| 39 | YTHDF3 | 4540632 4670717 7040609 | 3045 | 0.077 | 0.6268 | No | ||
| 40 | CAMK2A | 1740333 1940112 4150292 | 3239 | 0.064 | 0.6186 | No | ||
| 41 | ATE1 | 290491 6940722 | 3286 | 0.061 | 0.6182 | No | ||
| 42 | BAG5 | 2230368 6420537 | 3347 | 0.058 | 0.6170 | No | ||
| 43 | OLFML3 | 7100463 | 3516 | 0.049 | 0.6096 | No | ||
| 44 | CASK | 2340215 3290576 6770215 | 3677 | 0.041 | 0.6023 | No | ||
| 45 | THAP11 | 6940025 | 3733 | 0.038 | 0.6006 | No | ||
| 46 | CA2 | 1660113 1660600 | 3739 | 0.038 | 0.6017 | No | ||
| 47 | GALC | 2260685 4810161 6370040 | 4465 | 0.018 | 0.5631 | No | ||
| 48 | RTN1 | 4150452 4850176 | 4478 | 0.018 | 0.5630 | No | ||
| 49 | HOXA7 | 5910152 | 4545 | 0.017 | 0.5601 | No | ||
| 50 | NR4A3 | 2900021 5860095 5910039 | 4564 | 0.017 | 0.5597 | No | ||
| 51 | ERG | 50154 1770739 | 4736 | 0.014 | 0.5509 | No | ||
| 52 | HOXA2 | 2120121 | 5186 | 0.010 | 0.5270 | No | ||
| 53 | SH3BGRL2 | 1780239 | 5327 | 0.009 | 0.5197 | No | ||
| 54 | SEMA3A | 1780064 | 5367 | 0.009 | 0.5179 | No | ||
| 55 | CLDN5 | 6510717 | 5370 | 0.009 | 0.5181 | No | ||
| 56 | OPHN1 | 2360100 | 5454 | 0.008 | 0.5139 | No | ||
| 57 | ASB17 | 3520433 | 5469 | 0.008 | 0.5134 | No | ||
| 58 | LIN28 | 6590672 | 5528 | 0.008 | 0.5106 | No | ||
| 59 | NNT | 540253 1170471 5550092 6760397 | 5618 | 0.008 | 0.5060 | No | ||
| 60 | MYL3 | 6040563 | 5829 | 0.007 | 0.4949 | No | ||
| 61 | TNP1 | 3120048 | 5909 | 0.006 | 0.4908 | No | ||
| 62 | BSN | 5270347 | 6024 | 0.006 | 0.4849 | No | ||
| 63 | DLL1 | 1770377 | 6187 | 0.005 | 0.4763 | No | ||
| 64 | GRIN2B | 3800333 | 6310 | 0.005 | 0.4698 | No | ||
| 65 | CCNT2 | 2100390 4150563 | 6357 | 0.005 | 0.4675 | No | ||
| 66 | GSH1 | 2760242 | 6475 | 0.004 | 0.4613 | No | ||
| 67 | ARHGEF19 | 940102 | 6493 | 0.004 | 0.4605 | No | ||
| 68 | LIG1 | 1980438 4540112 5570609 | 6510 | 0.004 | 0.4598 | No | ||
| 69 | SLITRK4 | 5910022 | 6516 | 0.004 | 0.4597 | No | ||
| 70 | TSNAXIP1 | 1770270 6180577 | 6566 | 0.004 | 0.4572 | No | ||
| 71 | PLXNC1 | 5860315 | 6642 | 0.004 | 0.4532 | No | ||
| 72 | OMG | 6760066 | 6679 | 0.003 | 0.4514 | No | ||
| 73 | UCN2 | 4590372 | 6706 | 0.003 | 0.4501 | No | ||
| 74 | BAT4 | 2690128 6220451 | 6851 | 0.003 | 0.4424 | No | ||
| 75 | FSCN3 | 60093 | 7151 | 0.002 | 0.4263 | No | ||
| 76 | NNAT | 4920053 5130253 | 7203 | 0.002 | 0.4236 | No | ||
| 77 | MEIS1 | 1400575 | 7382 | 0.001 | 0.4140 | No | ||
| 78 | CFL2 | 380239 6200368 | 7416 | 0.001 | 0.4123 | No | ||
| 79 | ADAM22 | 5390368 | 7422 | 0.001 | 0.4121 | No | ||
| 80 | PVRL3 | 1400324 4200593 6650128 | 7613 | 0.001 | 0.4018 | No | ||
| 81 | FGF17 | 3130022 | 7964 | -0.000 | 0.3829 | No | ||
| 82 | FEZ1 | 2630725 4810324 | 8059 | -0.000 | 0.3778 | No | ||
| 83 | REST | 1500471 | 8229 | -0.001 | 0.3687 | No | ||
| 84 | MMP16 | 2680139 | 8347 | -0.001 | 0.3624 | No | ||
| 85 | PCDH8 | 2360047 4200452 | 8555 | -0.002 | 0.3512 | No | ||
| 86 | PCSK1N | 4010435 | 8683 | -0.002 | 0.3444 | No | ||
| 87 | PAX3 | 50551 | 8967 | -0.003 | 0.3292 | No | ||
| 88 | KIF21A | 2970338 | 9025 | -0.003 | 0.3262 | No | ||
| 89 | LAMA1 | 1770594 | 9333 | -0.004 | 0.3097 | No | ||
| 90 | FGF14 | 2630390 3520075 6770048 | 9385 | -0.004 | 0.3070 | No | ||
| 91 | FMR1 | 5050075 | 10007 | -0.006 | 0.2736 | No | ||
| 92 | CBX8 | 50671 | 10125 | -0.006 | 0.2675 | No | ||
| 93 | RAB3C | 5420072 | 10423 | -0.007 | 0.2517 | No | ||
| 94 | DPYSL3 | 1580594 | 10857 | -0.008 | 0.2285 | No | ||
| 95 | TRIB1 | 2320435 | 10950 | -0.009 | 0.2238 | No | ||
| 96 | PITX2 | 870537 2690139 | 11122 | -0.009 | 0.2149 | No | ||
| 97 | MYH2 | 2340433 | 11161 | -0.009 | 0.2132 | No | ||
| 98 | RFX4 | 3710100 5340215 | 11215 | -0.010 | 0.2106 | No | ||
| 99 | GPM6A | 1660044 2750152 | 11370 | -0.011 | 0.2027 | No | ||
| 100 | SMAD1 | 630537 1850333 | 11379 | -0.011 | 0.2026 | No | ||
| 101 | GJA12 | 430273 4560731 | 11847 | -0.013 | 0.1778 | No | ||
| 102 | NFIB | 460450 | 11897 | -0.013 | 0.1756 | No | ||
| 103 | IBSP | 2060048 | 12010 | -0.014 | 0.1700 | No | ||
| 104 | MB | 730019 | 12314 | -0.016 | 0.1542 | No | ||
| 105 | CA3 | 870687 5890390 | 12418 | -0.017 | 0.1492 | No | ||
| 106 | CSNK2B | 5080161 | 12666 | -0.019 | 0.1365 | No | ||
| 107 | CELSR3 | 5270731 7100082 | 12702 | -0.020 | 0.1353 | No | ||
| 108 | HOXC4 | 1940193 | 12720 | -0.020 | 0.1351 | No | ||
| 109 | CLDN17 | 3710672 | 12988 | -0.023 | 0.1214 | No | ||
| 110 | SCUBE2 | 2850315 5130040 | 13068 | -0.024 | 0.1179 | No | ||
| 111 | GRIK3 | 6380592 | 13451 | -0.029 | 0.0983 | No | ||
| 112 | PPP2R5C | 3310121 6650672 | 13814 | -0.035 | 0.0799 | No | ||
| 113 | HOXC6 | 3850133 4920736 | 13874 | -0.036 | 0.0780 | No | ||
| 114 | SLC36A3 | 430075 | 13925 | -0.037 | 0.0765 | No | ||
| 115 | RNF38 | 1940746 3140133 3840601 | 14056 | -0.040 | 0.0709 | No | ||
| 116 | ACRV1 | 4070161 | 14083 | -0.041 | 0.0709 | No | ||
| 117 | PRIMA1 | 1500673 6760671 | 14111 | -0.042 | 0.0709 | No | ||
| 118 | REC8L1 | 5900014 | 14246 | -0.045 | 0.0652 | No | ||
| 119 | FOXP2 | 3520561 4150372 4760524 | 14273 | -0.046 | 0.0654 | No | ||
| 120 | PRDM13 | 1430022 | 14278 | -0.046 | 0.0668 | No | ||
| 121 | BCL11A | 6860369 | 14495 | -0.052 | 0.0569 | No | ||
| 122 | CBLB | 3520465 4670348 | 14661 | -0.058 | 0.0500 | No | ||
| 123 | PYGO1 | 840575 | 14746 | -0.061 | 0.0476 | No | ||
| 124 | EXT1 | 4540603 | 14909 | -0.067 | 0.0411 | No | ||
| 125 | ETV4 | 2450605 | 15229 | -0.084 | 0.0268 | No | ||
| 126 | GRIA3 | 2900164 6130278 6860154 | 15238 | -0.085 | 0.0293 | No | ||
| 127 | EGR3 | 6940128 | 15260 | -0.086 | 0.0311 | No | ||
| 128 | TPM3 | 670600 3170296 5670167 6650471 | 15515 | -0.104 | 0.0210 | No | ||
| 129 | PDYN | 5720408 | 15587 | -0.112 | 0.0210 | No | ||
| 130 | IFRD2 | 4150110 | 15728 | -0.126 | 0.0178 | No | ||
| 131 | POFUT1 | 1570458 | 15773 | -0.129 | 0.0199 | No | ||
| 132 | NFIL3 | 4070377 | 16035 | -0.163 | 0.0114 | No | ||
| 133 | TOMM40 | 6110017 | 16432 | -0.219 | -0.0025 | No | ||
| 134 | SF4 | 3830341 | 16731 | -0.261 | -0.0096 | No | ||
| 135 | ITPR3 | 4010632 | 16780 | -0.270 | -0.0028 | No | ||
| 136 | TLE3 | 580040 4730121 | 17308 | -0.372 | -0.0185 | No | ||
| 137 | HSPB2 | 1990110 | 17335 | -0.378 | -0.0068 | No | ||
| 138 | OTUB1 | 610551 | 17456 | -0.412 | 0.0010 | No | ||
| 139 | CHD4 | 5420059 6130338 6380717 | 17466 | -0.415 | 0.0148 | No | ||
| 140 | RABAC1 | 2030435 2100632 | 17803 | -0.535 | 0.0152 | No | ||
| 141 | SIRT6 | 540731 1400735 | 18195 | -0.832 | 0.0228 | No |

