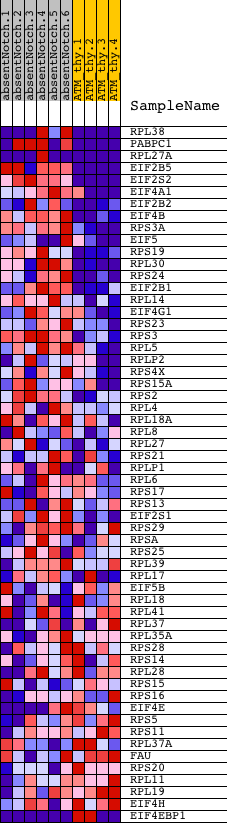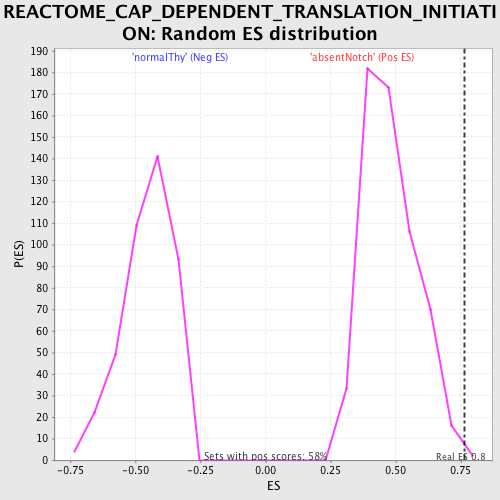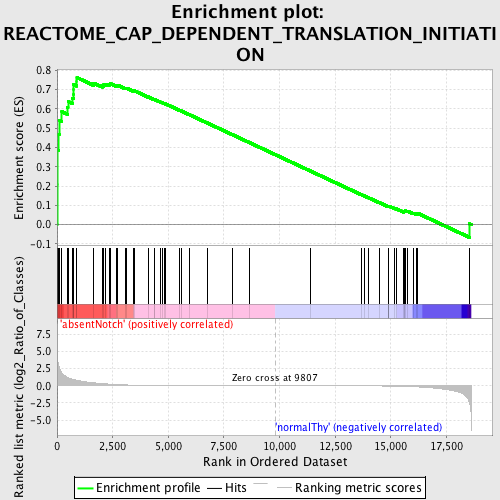
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy.phenotype_absentNotch_versus_normalThy.cls #absentNotch_versus_normalThy_repos |
| Phenotype | phenotype_absentNotch_versus_normalThy.cls#absentNotch_versus_normalThy_repos |
| Upregulated in class | absentNotch |
| GeneSet | REACTOME_CAP_DEPENDENT_TRANSLATION_INITIATION |
| Enrichment Score (ES) | 0.7628708 |
| Normalized Enrichment Score (NES) | 1.5960258 |
| Nominal p-value | 0.0034364262 |
| FDR q-value | 0.043758024 |
| FWER p-Value | 0.396 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RPL38 | 70040 4060138 6200601 | 2 | 7.446 | 0.2090 | Yes | ||
| 2 | PABPC1 | 2650180 2690253 6020632 1990270 | 3 | 6.314 | 0.3863 | Yes | ||
| 3 | RPL27A | 3130451 110195 6840494 | 72 | 2.999 | 0.4668 | Yes | ||
| 4 | EIF2B5 | 430315 6900400 | 92 | 2.667 | 0.5407 | Yes | ||
| 5 | EIF2S2 | 770095 2810487 | 214 | 1.852 | 0.5861 | Yes | ||
| 6 | EIF4A1 | 1990341 2810300 | 469 | 1.223 | 0.6068 | Yes | ||
| 7 | EIF2B2 | 4290048 | 490 | 1.174 | 0.6387 | Yes | ||
| 8 | EIF4B | 5390563 5270577 | 688 | 0.933 | 0.6543 | Yes | ||
| 9 | RPS3A | 2470086 | 731 | 0.899 | 0.6772 | Yes | ||
| 10 | EIF5 | 6380750 | 736 | 0.893 | 0.7021 | Yes | ||
| 11 | RPS19 | 5860066 | 739 | 0.891 | 0.7270 | Yes | ||
| 12 | RPL30 | 4810369 | 890 | 0.783 | 0.7409 | Yes | ||
| 13 | RPS24 | 5420377 | 892 | 0.783 | 0.7629 | Yes | ||
| 14 | EIF2B1 | 4610082 7040242 | 1642 | 0.423 | 0.7344 | No | ||
| 15 | RPL14 | 5080471 | 2026 | 0.312 | 0.7225 | No | ||
| 16 | EIF4G1 | 4070446 | 2101 | 0.290 | 0.7267 | No | ||
| 17 | RPS23 | 7100594 | 2190 | 0.268 | 0.7294 | No | ||
| 18 | RPS3 | 1570017 2370170 | 2339 | 0.234 | 0.7280 | No | ||
| 19 | RPL5 | 840095 | 2401 | 0.222 | 0.7310 | No | ||
| 20 | RPLP2 | 5290020 | 2669 | 0.174 | 0.7215 | No | ||
| 21 | RPS4X | 2690020 | 2720 | 0.166 | 0.7235 | No | ||
| 22 | RPS15A | 1170180 | 3079 | 0.120 | 0.7075 | No | ||
| 23 | RPS2 | 1340647 | 3130 | 0.114 | 0.7081 | No | ||
| 24 | RPL4 | 2940014 5420324 | 3429 | 0.087 | 0.6945 | No | ||
| 25 | RPL18A | 4570619 | 3479 | 0.083 | 0.6941 | No | ||
| 26 | RPL8 | 20296 | 4086 | 0.050 | 0.6629 | No | ||
| 27 | RPL27 | 1940088 | 4356 | 0.041 | 0.6496 | No | ||
| 28 | RPS21 | 3120673 | 4396 | 0.040 | 0.6486 | No | ||
| 29 | RPLP1 | 3140292 | 4652 | 0.032 | 0.6357 | No | ||
| 30 | RPL6 | 6040735 | 4715 | 0.031 | 0.6333 | No | ||
| 31 | RPS17 | 2360528 | 4804 | 0.029 | 0.6294 | No | ||
| 32 | RPS13 | 1570364 | 4892 | 0.028 | 0.6254 | No | ||
| 33 | EIF2S1 | 5360292 | 5509 | 0.019 | 0.5928 | No | ||
| 34 | RPS29 | 580324 | 5574 | 0.018 | 0.5898 | No | ||
| 35 | RPSA | 2360408 2680736 | 5956 | 0.015 | 0.5697 | No | ||
| 36 | RPS25 | 520139 | 6740 | 0.010 | 0.5278 | No | ||
| 37 | RPL39 | 2370446 | 7885 | 0.006 | 0.4663 | No | ||
| 38 | RPL17 | 2690167 5690504 | 8639 | 0.003 | 0.4259 | No | ||
| 39 | EIF5B | 1190102 4200440 | 11389 | -0.005 | 0.2779 | No | ||
| 40 | RPL18 | 6380181 3870195 | 13681 | -0.018 | 0.1550 | No | ||
| 41 | RPL41 | 6940112 | 13805 | -0.020 | 0.1489 | No | ||
| 42 | RPL37 | 3170494 3870750 4280494 | 13985 | -0.023 | 0.1399 | No | ||
| 43 | RPL35A | 2370706 | 14498 | -0.032 | 0.1132 | No | ||
| 44 | RPS28 | 3830221 | 14895 | -0.046 | 0.0932 | No | ||
| 45 | RPS14 | 430541 | 14898 | -0.046 | 0.0944 | No | ||
| 46 | RPL28 | 6900136 | 14904 | -0.047 | 0.0954 | No | ||
| 47 | RPS15 | 580100 | 14915 | -0.048 | 0.0962 | No | ||
| 48 | RPS16 | 510722 | 15142 | -0.061 | 0.0857 | No | ||
| 49 | EIF4E | 1580403 70133 6380215 | 15251 | -0.069 | 0.0818 | No | ||
| 50 | RPS5 | 580093 | 15580 | -0.099 | 0.0669 | No | ||
| 51 | RPS11 | 4120403 | 15606 | -0.101 | 0.0684 | No | ||
| 52 | RPL37A | 7040717 | 15622 | -0.102 | 0.0705 | No | ||
| 53 | FAU | 7100465 | 15664 | -0.107 | 0.0713 | No | ||
| 54 | RPS20 | 1740092 | 15738 | -0.114 | 0.0705 | No | ||
| 55 | RPL11 | 3780435 | 16011 | -0.144 | 0.0599 | No | ||
| 56 | RPL19 | 5550592 | 16174 | -0.168 | 0.0559 | No | ||
| 57 | EIF4H | 610324 | 16196 | -0.171 | 0.0596 | No | ||
| 58 | EIF4EBP1 | 60132 5720148 | 18539 | -2.520 | 0.0041 | No |