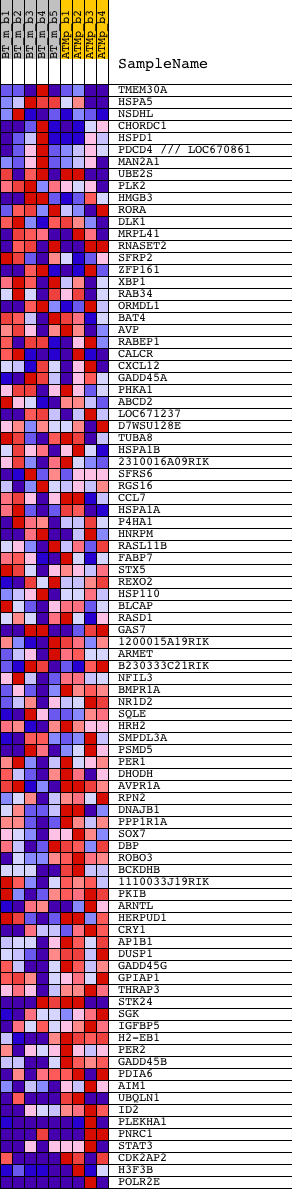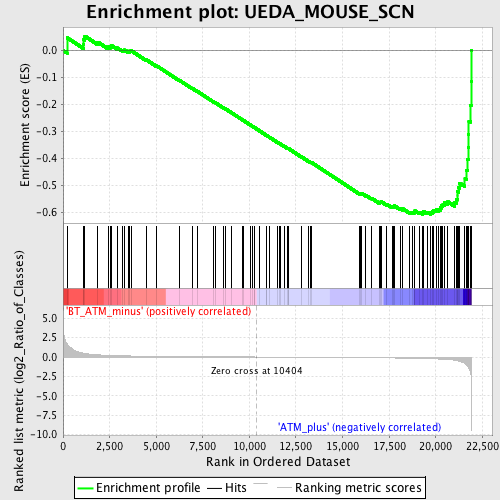
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | UEDA_MOUSE_SCN |
| Enrichment Score (ES) | -0.6086345 |
| Normalized Enrichment Score (NES) | -1.8354275 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.17096971 |
| FWER p-Value | 0.726 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TMEM30A | 1416446_at 1416447_at 1443619_at 1448339_at 1448340_at | 217 | 1.666 | 0.0470 | No | ||
| 2 | HSPA5 | 1416064_a_at 1427464_s_at 1447824_x_at | 1095 | 0.493 | 0.0237 | No | ||
| 3 | NSDHL | 1416222_at 1451799_at 1458831_at | 1110 | 0.487 | 0.0397 | No | ||
| 4 | CHORDC1 | 1435574_at 1460645_at | 1166 | 0.464 | 0.0531 | No | ||
| 5 | HSPD1 | 1426351_at | 1860 | 0.283 | 0.0310 | No | ||
| 6 | PDCD4 /// LOC670861 | 1418840_at | 2410 | 0.211 | 0.0131 | No | ||
| 7 | MAN2A1 | 1448647_at | 2522 | 0.200 | 0.0149 | No | ||
| 8 | UBE2S | 1416726_s_at 1430962_at | 2607 | 0.190 | 0.0175 | No | ||
| 9 | PLK2 | 1427005_at | 2914 | 0.167 | 0.0092 | No | ||
| 10 | HMGB3 | 1416155_at | 3204 | 0.150 | 0.0011 | No | ||
| 11 | RORA | 1420583_a_at 1424034_at 1424035_at 1436325_at 1436326_at 1441085_at 1443511_at 1443647_at 1446360_at 1455165_at 1457177_at 1458129_at | 3298 | 0.145 | 0.0018 | No | ||
| 12 | DLK1 | 1449939_s_at | 3496 | 0.136 | -0.0026 | No | ||
| 13 | MRPL41 | 1428588_a_at 1428589_at 1428590_at | 3583 | 0.132 | -0.0020 | No | ||
| 14 | RNASET2 | 1431785_at 1452734_at 1456012_x_at | 3658 | 0.129 | -0.0010 | No | ||
| 15 | SFRP2 | 1448201_at | 4455 | 0.101 | -0.0340 | No | ||
| 16 | ZFP161 | 1420864_at 1420865_at 1420866_at 1437585_x_at | 5032 | 0.086 | -0.0574 | No | ||
| 17 | XBP1 | 1420011_s_at 1420012_at 1420886_a_at 1437223_s_at | 6223 | 0.062 | -0.1098 | No | ||
| 18 | RAB34 | 1416590_a_at 1416591_at | 6948 | 0.049 | -0.1413 | No | ||
| 19 | ORMDL1 | 1451219_at | 7221 | 0.044 | -0.1522 | No | ||
| 20 | BAT4 | 1420284_at 1423163_at | 8089 | 0.031 | -0.1908 | No | ||
| 21 | AVP | 1450794_at | 8166 | 0.030 | -0.1933 | No | ||
| 22 | RABEP1 | 1421305_x_at 1426023_a_at 1427448_at 1443856_at 1444930_at 1459494_at | 8593 | 0.024 | -0.2119 | No | ||
| 23 | CALCR | 1418688_at | 8720 | 0.022 | -0.2169 | No | ||
| 24 | CXCL12 | 1417574_at 1439084_at 1448823_at | 9049 | 0.018 | -0.2313 | No | ||
| 25 | GADD45A | 1449519_at | 9649 | 0.010 | -0.2584 | No | ||
| 26 | PHKA1 | 1422743_at 1422744_at 1435567_at | 9679 | 0.010 | -0.2594 | No | ||
| 27 | ABCD2 | 1419748_at 1438431_at 1439835_x_at 1446962_at 1456812_at | 10088 | 0.004 | -0.2779 | No | ||
| 28 | LOC671237 | 1422660_at | 10153 | 0.003 | -0.2807 | No | ||
| 29 | D7WSU128E | 1418132_a_at 1434078_at 1448968_at 1456217_at | 10252 | 0.002 | -0.2852 | No | ||
| 30 | TUBA8 | 1419518_at | 10283 | 0.002 | -0.2865 | No | ||
| 31 | HSPA1B | 1427126_at 1427127_x_at 1452318_a_at | 10556 | -0.002 | -0.2989 | No | ||
| 32 | 2310016A09RIK | 1451322_at | 10895 | -0.007 | -0.3141 | No | ||
| 33 | SFRS6 | 1416720_at 1416721_s_at 1447898_s_at 1448454_at | 10916 | -0.007 | -0.3148 | No | ||
| 34 | RGS16 | 1426037_a_at 1451452_a_at 1455265_a_at | 11068 | -0.009 | -0.3214 | No | ||
| 35 | CCL7 | 1421228_at | 11527 | -0.015 | -0.3419 | No | ||
| 36 | HSPA1A | 1452388_at | 11604 | -0.016 | -0.3448 | No | ||
| 37 | P4HA1 | 1426519_at 1452094_at | 11674 | -0.017 | -0.3474 | No | ||
| 38 | HNRPM | 1426698_a_at | 11900 | -0.019 | -0.3571 | No | ||
| 39 | RASL11B | 1423854_a_at | 12032 | -0.021 | -0.3623 | No | ||
| 40 | FABP7 | 1450779_at | 12079 | -0.021 | -0.3637 | No | ||
| 41 | STX5 | 1419952_at 1448267_at 1449679_s_at | 12084 | -0.022 | -0.3632 | No | ||
| 42 | REXO2 | 1451259_at | 12805 | -0.031 | -0.3950 | No | ||
| 43 | HSP110 | 1423566_a_at 1425993_a_at | 13193 | -0.037 | -0.4115 | No | ||
| 44 | BLCAP | 1420631_a_at | 13290 | -0.038 | -0.4146 | No | ||
| 45 | RASD1 | 1423619_at | 13358 | -0.039 | -0.4163 | No | ||
| 46 | GAS7 | 1417859_at 1431400_a_at 1445685_at | 15917 | -0.081 | -0.5307 | No | ||
| 47 | 1200015A19RIK | 1451172_at | 15950 | -0.082 | -0.5293 | No | ||
| 48 | ARMET | 1428112_at 1459455_at | 16007 | -0.083 | -0.5291 | No | ||
| 49 | B230333C21RIK | 1457265_at 1457686_at | 16256 | -0.087 | -0.5374 | No | ||
| 50 | NFIL3 | 1418932_at | 16559 | -0.094 | -0.5480 | No | ||
| 51 | BMPR1A | 1425491_at 1425492_at 1425493_at 1425494_s_at 1445413_at 1451729_at | 16993 | -0.105 | -0.5643 | No | ||
| 52 | NR1D2 | 1416958_at 1416959_at 1457250_x_at | 17030 | -0.106 | -0.5623 | No | ||
| 53 | SQLE | 1415993_at | 17071 | -0.107 | -0.5605 | No | ||
| 54 | HRH2 | 1423639_at | 17389 | -0.115 | -0.5710 | No | ||
| 55 | SMPDL3A | 1416635_at | 17670 | -0.123 | -0.5797 | No | ||
| 56 | PSMD5 | 1423234_at | 17740 | -0.125 | -0.5785 | No | ||
| 57 | PER1 | 1449851_at | 17792 | -0.126 | -0.5766 | No | ||
| 58 | DHODH | 1417581_at 1417582_s_at | 18120 | -0.136 | -0.5869 | No | ||
| 59 | AVPR1A | 1418603_at 1418604_at | 18247 | -0.140 | -0.5878 | No | ||
| 60 | RPN2 | 1418896_a_at | 18621 | -0.154 | -0.5997 | No | ||
| 61 | DNAJB1 | 1416755_at 1416756_at | 18767 | -0.159 | -0.6009 | No | ||
| 62 | PPP1R1A | 1422605_at | 18847 | -0.163 | -0.5989 | No | ||
| 63 | SOX7 | 1416564_at 1432495_at | 18894 | -0.165 | -0.5954 | No | ||
| 64 | DBP | 1418174_at 1438211_s_at | 19121 | -0.175 | -0.5997 | No | ||
| 65 | ROBO3 | 1421565_at 1436634_at | 19305 | -0.185 | -0.6018 | Yes | ||
| 66 | BCKDHB | 1427153_at | 19358 | -0.188 | -0.5977 | Yes | ||
| 67 | 1110033J19RIK | 1452730_at | 19556 | -0.200 | -0.5999 | Yes | ||
| 68 | PKIB | 1421137_a_at 1421138_a_at | 19748 | -0.214 | -0.6013 | Yes | ||
| 69 | ARNTL | 1425099_a_at 1446544_at 1457740_at | 19829 | -0.220 | -0.5975 | Yes | ||
| 70 | HERPUD1 | 1435626_a_at 1448185_at | 19880 | -0.225 | -0.5920 | Yes | ||
| 71 | CRY1 | 1433733_a_at | 20024 | -0.239 | -0.5904 | Yes | ||
| 72 | AP1B1 | 1416279_at | 20179 | -0.257 | -0.5887 | Yes | ||
| 73 | DUSP1 | 1448830_at | 20263 | -0.266 | -0.5834 | Yes | ||
| 74 | GADD45G | 1453851_a_at | 20300 | -0.269 | -0.5759 | Yes | ||
| 75 | GPIAP1 | 1416461_at 1416462_at 1416463_at 1433901_at 1437162_at 1448347_a_at 1448348_at | 20397 | -0.283 | -0.5706 | Yes | ||
| 76 | THRAP3 | 1427408_a_at 1438286_at 1446031_at 1452125_at 1460545_at | 20478 | -0.293 | -0.5642 | Yes | ||
| 77 | STK24 | 1426247_at 1426248_at 1440420_at 1446107_at | 20662 | -0.322 | -0.5616 | Yes | ||
| 78 | SGK | 1416041_at | 21030 | -0.417 | -0.5642 | Yes | ||
| 79 | IGFBP5 | 1422313_a_at 1452114_s_at | 21129 | -0.455 | -0.5531 | Yes | ||
| 80 | H2-EB1 | 1417025_at | 21171 | -0.470 | -0.5389 | Yes | ||
| 81 | PER2 | 1417602_at 1417603_at 1445892_at 1457350_at | 21172 | -0.470 | -0.5228 | Yes | ||
| 82 | GADD45B | 1420197_at 1449773_s_at 1450971_at | 21244 | -0.512 | -0.5085 | Yes | ||
| 83 | PDIA6 | 1423648_at 1430983_at | 21294 | -0.547 | -0.4921 | Yes | ||
| 84 | AIM1 | 1426942_at 1444412_at | 21580 | -0.844 | -0.4762 | Yes | ||
| 85 | UBQLN1 | 1419385_a_at 1424368_s_at 1450345_at | 21677 | -1.042 | -0.4450 | Yes | ||
| 86 | ID2 | 1422537_a_at 1435176_a_at 1453596_at | 21727 | -1.235 | -0.4050 | Yes | ||
| 87 | PLEKHA1 | 1417965_at 1443111_at | 21759 | -1.350 | -0.3603 | Yes | ||
| 88 | PNRC1 | 1433668_at 1438524_x_at 1439513_at | 21780 | -1.414 | -0.3129 | Yes | ||
| 89 | STAT3 | 1424272_at 1426587_a_at 1459961_a_at 1460700_at | 21793 | -1.500 | -0.2621 | Yes | ||
| 90 | CDK2AP2 | 1417161_at 1456490_at | 21875 | -1.812 | -0.2039 | Yes | ||
| 91 | H3F3B | 1420376_a_at 1430357_at 1455725_a_at | 21913 | -2.628 | -0.1157 | Yes | ||
| 92 | POLR2E | 1417138_s_at 1451093_at 1458326_at | 21922 | -3.411 | 0.0005 | Yes |