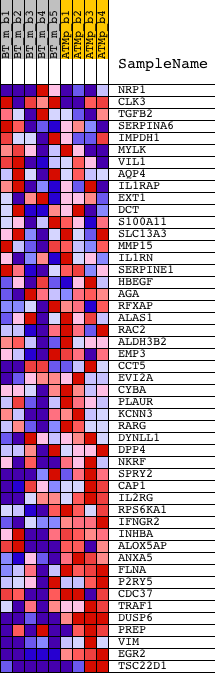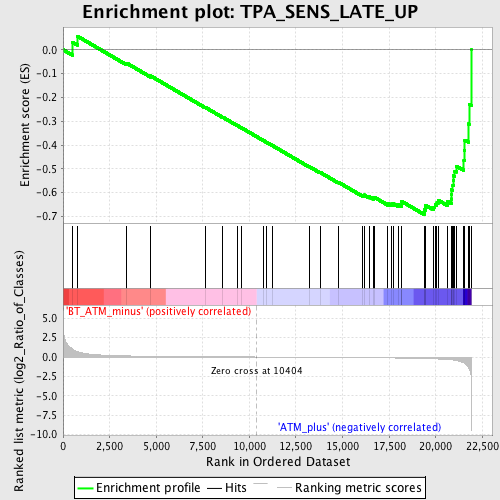
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | TPA_SENS_LATE_UP |
| Enrichment Score (ES) | -0.6916465 |
| Normalized Enrichment Score (NES) | -1.8848548 |
| Nominal p-value | 0.002793296 |
| FDR q-value | 0.15760513 |
| FWER p-Value | 0.493 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NRP1 | 1418084_at 1448943_at 1448944_at 1457198_at | 508 | 0.999 | 0.0313 | No | ||
| 2 | CLK3 | 1423849_a_at 1441472_at | 763 | 0.690 | 0.0573 | No | ||
| 3 | TGFB2 | 1423250_a_at 1438303_at 1446141_at 1450922_a_at 1450923_at | 3404 | 0.140 | -0.0557 | No | ||
| 4 | SERPINA6 | 1448506_at | 4699 | 0.094 | -0.1097 | No | ||
| 5 | IMPDH1 | 1423239_at | 7635 | 0.038 | -0.2417 | No | ||
| 6 | MYLK | 1425504_at 1425505_at 1425506_at | 8585 | 0.024 | -0.2837 | No | ||
| 7 | VIL1 | 1448837_at | 9365 | 0.014 | -0.3186 | No | ||
| 8 | AQP4 | 1425382_a_at 1434449_at 1447745_at | 9570 | 0.011 | -0.3273 | No | ||
| 9 | IL1RAP | 1421843_at 1439697_at 1442614_at 1449585_at | 10761 | -0.004 | -0.3814 | No | ||
| 10 | EXT1 | 1417730_at 1439877_at 1440011_at 1440092_at 1456655_at 1458054_at 1458296_at | 10901 | -0.007 | -0.3874 | No | ||
| 11 | DCT | 1418028_at | 11232 | -0.011 | -0.4019 | No | ||
| 12 | S100A11 | 1460351_at | 13207 | -0.037 | -0.4901 | No | ||
| 13 | SLC13A3 | 1416560_at 1438377_x_at 1439384_at 1439385_x_at | 13802 | -0.046 | -0.5147 | No | ||
| 14 | MMP15 | 1422597_at 1437462_x_at 1439734_at | 14782 | -0.061 | -0.5561 | No | ||
| 15 | IL1RN | 1423017_a_at 1425663_at 1451798_at | 16076 | -0.084 | -0.6106 | No | ||
| 16 | SERPINE1 | 1419149_at | 16158 | -0.085 | -0.6097 | No | ||
| 17 | HBEGF | 1418349_at 1418350_at | 16439 | -0.092 | -0.6175 | No | ||
| 18 | AGA | 1434665_at | 16652 | -0.096 | -0.6219 | No | ||
| 19 | RFXAP | 1418859_at | 16707 | -0.098 | -0.6190 | No | ||
| 20 | ALAS1 | 1424126_at 1442331_at 1455282_x_at | 17439 | -0.117 | -0.6461 | No | ||
| 21 | RAC2 | 1417620_at 1440208_at | 17615 | -0.121 | -0.6474 | No | ||
| 22 | ALDH3B2 | 1456317_at | 17739 | -0.125 | -0.6463 | No | ||
| 23 | EMP3 | 1417104_at | 17994 | -0.133 | -0.6506 | No | ||
| 24 | CCT5 | 1417258_at | 18164 | -0.138 | -0.6508 | No | ||
| 25 | EVI2A | 1443894_at 1450241_a_at 1458788_at 1460055_at | 18169 | -0.138 | -0.6435 | No | ||
| 26 | CYBA | 1454268_a_at | 18170 | -0.138 | -0.6360 | No | ||
| 27 | PLAUR | 1452521_a_at | 19389 | -0.190 | -0.6813 | Yes | ||
| 28 | KCNN3 | 1421632_at 1459308_at | 19418 | -0.191 | -0.6722 | Yes | ||
| 29 | RARG | 1419415_a_at 1419416_a_at | 19450 | -0.194 | -0.6630 | Yes | ||
| 30 | DYNLL1 | 1448682_at | 19471 | -0.195 | -0.6533 | Yes | ||
| 31 | DPP4 | 1416697_at 1438279_at 1441242_at 1441342_at 1459973_x_at | 19898 | -0.228 | -0.6603 | Yes | ||
| 32 | NKRF | 1434398_at 1441325_at 1445983_at | 19970 | -0.233 | -0.6509 | Yes | ||
| 33 | SPRY2 | 1421656_at 1436584_at | 20076 | -0.244 | -0.6424 | Yes | ||
| 34 | CAP1 | 1417461_at 1417462_at | 20136 | -0.252 | -0.6313 | Yes | ||
| 35 | IL2RG | 1416295_a_at 1416296_at | 20630 | -0.317 | -0.6366 | Yes | ||
| 36 | RPS6KA1 | 1416896_at | 20840 | -0.358 | -0.6266 | Yes | ||
| 37 | IFNGR2 | 1423557_at 1423558_at | 20850 | -0.361 | -0.6073 | Yes | ||
| 38 | INHBA | 1422053_at 1458291_at | 20861 | -0.364 | -0.5880 | Yes | ||
| 39 | ALOX5AP | 1452016_at 1459234_at | 20929 | -0.383 | -0.5702 | Yes | ||
| 40 | ANXA5 | 1425567_a_at | 20950 | -0.387 | -0.5500 | Yes | ||
| 41 | FLNA | 1426677_at | 20973 | -0.396 | -0.5294 | Yes | ||
| 42 | P2RY5 | 1428615_at 1444627_at | 21037 | -0.420 | -0.5094 | Yes | ||
| 43 | CDC37 | 1416819_at 1416820_at | 21126 | -0.454 | -0.4887 | Yes | ||
| 44 | TRAF1 | 1423602_at 1445452_at | 21524 | -0.759 | -0.4654 | Yes | ||
| 45 | DUSP6 | 1415834_at | 21545 | -0.791 | -0.4232 | Yes | ||
| 46 | PREP | 1422796_at 1436198_at 1439942_at | 21574 | -0.826 | -0.3795 | Yes | ||
| 47 | VIM | 1438118_x_at 1450641_at 1456292_a_at | 21782 | -1.421 | -0.3114 | Yes | ||
| 48 | EGR2 | 1427682_a_at 1427683_at | 21804 | -1.550 | -0.2279 | Yes | ||
| 49 | TSC22D1 | 1425742_a_at 1433899_x_at 1447360_at 1454758_a_at 1454971_x_at 1456132_x_at | 21928 | -4.288 | 0.0003 | Yes |