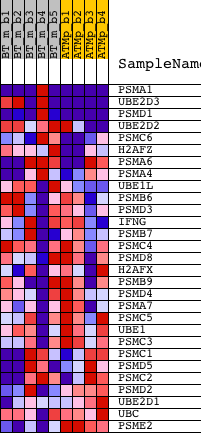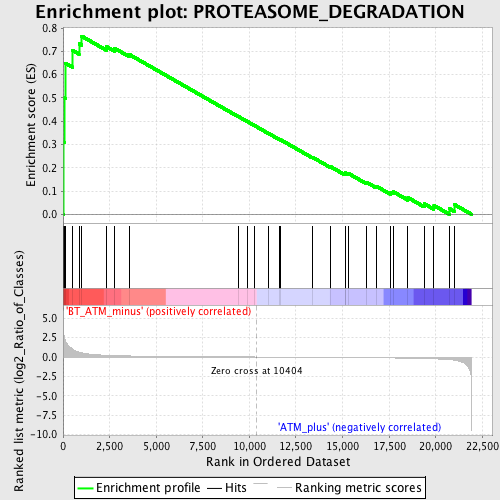
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | PROTEASOME_DEGRADATION |
| Enrichment Score (ES) | 0.7651497 |
| Normalized Enrichment Score (NES) | 1.7407305 |
| Nominal p-value | 0.0018115942 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.792 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PSMA1 | 1415695_at 1436769_at 1436770_x_at 1458292_at | 13 | 4.351 | 0.3114 | Yes | ||
| 2 | UBE2D3 | 1423112_at 1423113_a_at 1423114_at 1450858_a_at 1450859_s_at 1455479_a_at 1455480_s_at | 58 | 2.672 | 0.5009 | Yes | ||
| 3 | PSMD1 | 1419799_at 1429291_at | 120 | 2.105 | 0.6490 | Yes | ||
| 4 | UBE2D2 | 1416475_at 1416476_a_at 1416477_at 1439655_at 1445885_at 1448356_at | 499 | 1.012 | 0.7043 | Yes | ||
| 5 | PSMC6 | 1417771_a_at | 859 | 0.620 | 0.7324 | Yes | ||
| 6 | H2AFZ | 1441370_at | 992 | 0.541 | 0.7651 | Yes | ||
| 7 | PSMA6 | 1416506_at 1435316_at 1435317_x_at 1437144_x_at | 2338 | 0.220 | 0.7195 | No | ||
| 8 | PSMA4 | 1460339_at | 2762 | 0.177 | 0.7129 | No | ||
| 9 | UBE1L | 1426971_at 1437317_at | 3553 | 0.133 | 0.6864 | No | ||
| 10 | PSMB6 | 1448822_at | 9419 | 0.013 | 0.4196 | No | ||
| 11 | PSMD3 | 1448479_at | 9918 | 0.006 | 0.3973 | No | ||
| 12 | IFNG | 1425947_at | 10255 | 0.002 | 0.3822 | No | ||
| 13 | PSMB7 | 1416240_at | 11052 | -0.009 | 0.3464 | No | ||
| 14 | PSMC4 | 1416290_a_at 1416291_at | 11602 | -0.016 | 0.3225 | No | ||
| 15 | PSMD8 | 1423296_at | 11687 | -0.017 | 0.3199 | No | ||
| 16 | H2AFX | 1416746_at | 13384 | -0.040 | 0.2453 | No | ||
| 17 | PSMB9 | 1450696_at | 14333 | -0.054 | 0.2059 | No | ||
| 18 | PSMD4 | 1418874_a_at 1425859_a_at 1451725_a_at | 15139 | -0.067 | 0.1739 | No | ||
| 19 | PSMA7 | 1423567_a_at 1423568_at | 15155 | -0.067 | 0.1781 | No | ||
| 20 | PSMC5 | 1415740_at | 15323 | -0.070 | 0.1755 | No | ||
| 21 | UBE1 | 1442767_s_at 1448116_at | 16314 | -0.089 | 0.1367 | No | ||
| 22 | PSMC3 | 1416282_at | 16809 | -0.100 | 0.1213 | No | ||
| 23 | PSMC1 | 1416005_at | 17592 | -0.121 | 0.0943 | No | ||
| 24 | PSMD5 | 1423234_at | 17740 | -0.125 | 0.0965 | No | ||
| 25 | PSMC2 | 1426611_at 1435859_x_at 1443307_at | 18517 | -0.150 | 0.0718 | No | ||
| 26 | PSMD2 | 1415831_at | 19394 | -0.190 | 0.0455 | No | ||
| 27 | UBE2D1 | 1424062_at | 19913 | -0.229 | 0.0382 | No | ||
| 28 | UBC | 1420494_x_at 1425965_at 1425966_x_at 1432827_x_at 1437666_x_at 1454373_x_at | 20757 | -0.338 | 0.0240 | No | ||
| 29 | PSME2 | 1417189_at | 21023 | -0.414 | 0.0416 | No |