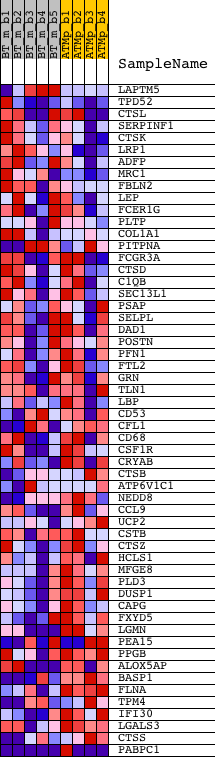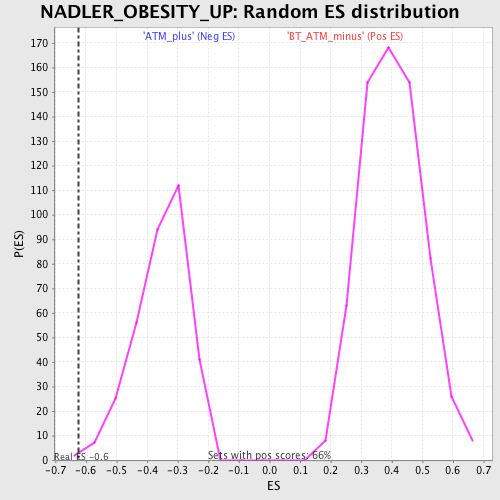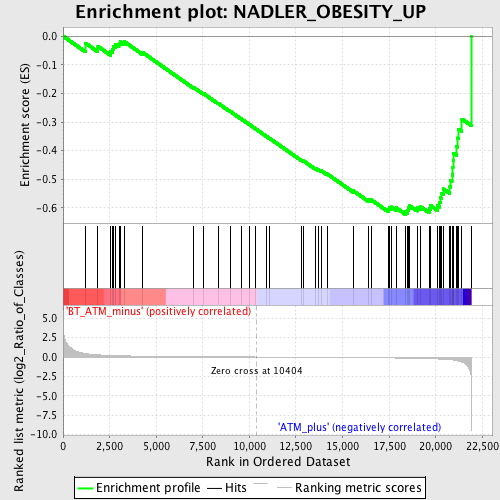
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | NADLER_OBESITY_UP |
| Enrichment Score (ES) | -0.6228424 |
| Normalized Enrichment Score (NES) | -1.7716966 |
| Nominal p-value | 0.005934718 |
| FDR q-value | 0.16883628 |
| FWER p-Value | 0.946 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | LAPTM5 | 1426025_s_at 1436905_x_at 1447742_at 1459841_x_at | 1195 | 0.456 | -0.0239 | No | ||
| 2 | TPD52 | 1419493_a_at 1419494_a_at 1436008_at 1458668_at | 1869 | 0.281 | -0.0357 | No | ||
| 3 | CTSL | 1451310_a_at 1457724_at | 2542 | 0.197 | -0.0532 | No | ||
| 4 | SERPINF1 | 1416168_at 1453724_a_at | 2646 | 0.187 | -0.0453 | No | ||
| 5 | CTSK | 1450652_at | 2711 | 0.181 | -0.0360 | No | ||
| 6 | LRP1 | 1442849_at 1448655_at | 2823 | 0.174 | -0.0294 | No | ||
| 7 | ADFP | 1448318_at | 3002 | 0.162 | -0.0266 | No | ||
| 8 | MRC1 | 1450430_at | 3070 | 0.157 | -0.0191 | No | ||
| 9 | FBLN2 | 1423407_a_at 1438375_at | 3276 | 0.146 | -0.0187 | No | ||
| 10 | LEP | 1422582_at | 4282 | 0.106 | -0.0575 | No | ||
| 11 | FCER1G | 1418340_at | 7011 | 0.048 | -0.1789 | No | ||
| 12 | PLTP | 1417963_at 1456424_s_at | 7563 | 0.039 | -0.2014 | No | ||
| 13 | COL1A1 | 1423669_at 1455494_at | 8317 | 0.028 | -0.2340 | No | ||
| 14 | PITPNA | 1423282_at 1423283_at | 8992 | 0.019 | -0.2635 | No | ||
| 15 | FCGR3A | 1425225_at 1448620_at | 9554 | 0.011 | -0.2884 | No | ||
| 16 | CTSD | 1448118_a_at | 10030 | 0.005 | -0.3097 | No | ||
| 17 | C1QB | 1417063_at 1434366_x_at 1437726_x_at 1457322_at | 10329 | 0.001 | -0.3233 | No | ||
| 18 | SEC13L1 | 1416241_at | 10924 | -0.007 | -0.3500 | No | ||
| 19 | PSAP | 1415687_a_at 1421813_a_at | 11097 | -0.009 | -0.3572 | No | ||
| 20 | SELPL | 1449127_at | 12821 | -0.032 | -0.4339 | No | ||
| 21 | DAD1 | 1418528_a_at 1454860_x_at | 12884 | -0.032 | -0.4345 | No | ||
| 22 | POSTN | 1423606_at | 13571 | -0.042 | -0.4630 | No | ||
| 23 | PFN1 | 1449018_at | 13688 | -0.044 | -0.4653 | No | ||
| 24 | FTL2 | 1422301_at | 13870 | -0.047 | -0.4704 | No | ||
| 25 | GRN | 1438629_x_at 1448148_at 1456567_x_at | 14184 | -0.052 | -0.4813 | No | ||
| 26 | TLN1 | 1436042_at 1448402_at 1457782_at | 15572 | -0.075 | -0.5396 | No | ||
| 27 | LBP | 1448550_at | 16409 | -0.091 | -0.5717 | No | ||
| 28 | CD53 | 1439589_at 1448617_at 1459744_at | 16542 | -0.094 | -0.5714 | No | ||
| 29 | CFL1 | 1448346_at 1455138_x_at | 17478 | -0.118 | -0.6062 | No | ||
| 30 | CD68 | 1449164_at | 17501 | -0.118 | -0.5992 | No | ||
| 31 | CSF1R | 1419872_at 1419873_s_at 1423593_a_at | 17623 | -0.122 | -0.5966 | No | ||
| 32 | CRYAB | 1416455_a_at 1434369_a_at 1435837_at | 17881 | -0.129 | -0.5996 | No | ||
| 33 | CTSB | 1417490_at 1417491_at 1417492_at 1444987_at 1448732_at | 18390 | -0.145 | -0.6131 | Yes | ||
| 34 | ATP6V1C1 | 1419544_at 1419545_a_at 1419546_at | 18497 | -0.149 | -0.6079 | Yes | ||
| 35 | NEDD8 | 1423715_a_at 1440996_at 1445811_at | 18540 | -0.151 | -0.5996 | Yes | ||
| 36 | CCL9 | 1417936_at 1448898_at | 18611 | -0.153 | -0.5925 | Yes | ||
| 37 | UCP2 | 1448188_at 1459740_s_at 1459741_x_at | 19010 | -0.170 | -0.5993 | Yes | ||
| 38 | CSTB | 1422506_a_at 1422507_at | 19182 | -0.178 | -0.5951 | Yes | ||
| 39 | CTSZ | 1417868_a_at 1417869_s_at 1417870_x_at | 19655 | -0.207 | -0.6027 | Yes | ||
| 40 | HCLS1 | 1418842_at | 19728 | -0.213 | -0.5916 | Yes | ||
| 41 | MFGE8 | 1420911_a_at | 20088 | -0.245 | -0.5915 | Yes | ||
| 42 | PLD3 | 1416013_at | 20208 | -0.260 | -0.5795 | Yes | ||
| 43 | DUSP1 | 1448830_at | 20263 | -0.266 | -0.5641 | Yes | ||
| 44 | CAPG | 1447803_x_at 1450355_a_at | 20328 | -0.273 | -0.5486 | Yes | ||
| 45 | FXYD5 | 1418296_at | 20425 | -0.287 | -0.5337 | Yes | ||
| 46 | LGMN | 1448883_at | 20771 | -0.342 | -0.5264 | Yes | ||
| 47 | PEA15 | 1416406_at 1416407_at 1459733_at | 20796 | -0.346 | -0.5042 | Yes | ||
| 48 | PPGB | 1445659_at 1448128_at 1458466_at | 20885 | -0.370 | -0.4832 | Yes | ||
| 49 | ALOX5AP | 1452016_at 1459234_at | 20929 | -0.383 | -0.4594 | Yes | ||
| 50 | BASP1 | 1428572_at | 20939 | -0.384 | -0.4340 | Yes | ||
| 51 | FLNA | 1426677_at | 20973 | -0.396 | -0.4088 | Yes | ||
| 52 | TPM4 | 1433883_at | 21123 | -0.452 | -0.3852 | Yes | ||
| 53 | IFI30 | 1422476_at | 21168 | -0.469 | -0.3556 | Yes | ||
| 54 | LGALS3 | 1426808_at 1445626_at | 21226 | -0.499 | -0.3246 | Yes | ||
| 55 | CTSS | 1448591_at | 21416 | -0.652 | -0.2894 | Yes | ||
| 56 | PABPC1 | 1418883_a_at 1419500_at 1453840_at | 21929 | -4.646 | 0.0002 | Yes |