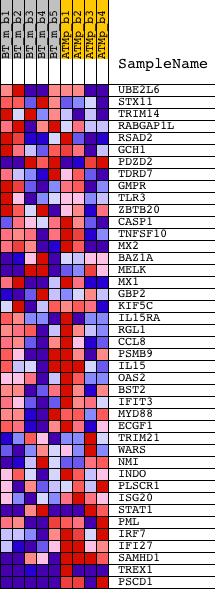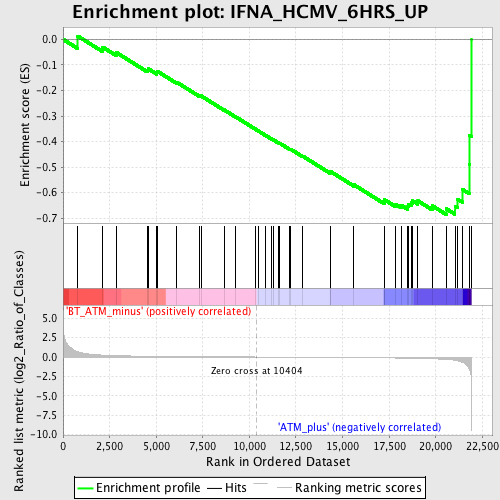
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | IFNA_HCMV_6HRS_UP |
| Enrichment Score (ES) | -0.6851795 |
| Normalized Enrichment Score (NES) | -1.8066735 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.14645763 |
| FWER p-Value | 0.84 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | UBE2L6 | 1417172_at 1447633_x_at | 790 | 0.668 | 0.0127 | No | ||
| 2 | STX11 | 1423576_a_at 1451032_at 1453228_at 1457780_at | 2124 | 0.245 | -0.0303 | No | ||
| 3 | TRIM14 | 1430796_at | 2866 | 0.170 | -0.0517 | No | ||
| 4 | RABGAP1L | 1421488_at 1429196_at 1429197_s_at 1434062_at 1441202_at 1453365_at 1457178_at 1458155_at | 4522 | 0.099 | -0.1201 | No | ||
| 5 | RSAD2 | 1421008_at 1421009_at 1436058_at | 4577 | 0.098 | -0.1154 | No | ||
| 6 | GCH1 | 1420499_at 1429692_s_at | 5036 | 0.086 | -0.1300 | No | ||
| 7 | PDZD2 | 1435553_at 1443618_at 1445203_at 1453896_at | 5052 | 0.086 | -0.1244 | No | ||
| 8 | TDRD7 | 1426716_at 1459216_at | 6114 | 0.064 | -0.1682 | No | ||
| 9 | GMPR | 1448530_at | 7340 | 0.042 | -0.2211 | No | ||
| 10 | TLR3 | 1422781_at 1422782_s_at | 7404 | 0.042 | -0.2209 | No | ||
| 11 | ZBTB20 | 1422064_a_at 1437065_at 1437066_at 1437598_at 1438443_at 1439128_at 1439278_at 1440565_at 1443471_at 1451577_at | 8645 | 0.024 | -0.2758 | No | ||
| 12 | CASP1 | 1449265_at | 9262 | 0.015 | -0.3028 | No | ||
| 13 | TNFSF10 | 1420412_at 1439680_at 1459913_at | 10357 | 0.001 | -0.3528 | No | ||
| 14 | MX2 | 1419676_at | 10480 | -0.001 | -0.3583 | No | ||
| 15 | BAZ1A | 1433599_at 1447930_at | 10860 | -0.006 | -0.3751 | No | ||
| 16 | MELK | 1416558_at | 11206 | -0.011 | -0.3901 | No | ||
| 17 | MX1 | 1451905_a_at | 11317 | -0.012 | -0.3943 | No | ||
| 18 | GBP2 | 1418240_at 1435906_x_at | 11561 | -0.015 | -0.4043 | No | ||
| 19 | KIF5C | 1422945_a_at 1427635_at 1447454_at 1450804_at 1455266_at 1457255_x_at | 11629 | -0.016 | -0.4062 | No | ||
| 20 | IL15RA | 1422397_a_at 1448681_at | 12168 | -0.023 | -0.4291 | No | ||
| 21 | RGL1 | 1418535_at 1440662_at 1449124_at | 12222 | -0.024 | -0.4298 | No | ||
| 22 | CCL8 | 1419684_at | 12872 | -0.032 | -0.4571 | No | ||
| 23 | PSMB9 | 1450696_at | 14333 | -0.054 | -0.5198 | No | ||
| 24 | IL15 | 1418219_at | 14363 | -0.055 | -0.5171 | No | ||
| 25 | OAS2 | 1425065_at | 15618 | -0.075 | -0.5689 | No | ||
| 26 | BST2 | 1424921_at | 17242 | -0.111 | -0.6349 | No | ||
| 27 | IFIT3 | 1449025_at | 17266 | -0.112 | -0.6278 | No | ||
| 28 | MYD88 | 1419272_at | 17870 | -0.129 | -0.6459 | No | ||
| 29 | ECGF1 | 1432181_s_at | 18156 | -0.138 | -0.6489 | No | ||
| 30 | TRIM21 | 1418077_at 1448940_at | 18512 | -0.150 | -0.6541 | No | ||
| 31 | WARS | 1415694_at 1425106_a_at 1434813_x_at 1437832_x_at | 18548 | -0.151 | -0.6447 | No | ||
| 32 | NMI | 1425719_a_at | 18694 | -0.156 | -0.6399 | No | ||
| 33 | INDO | 1420437_at | 18772 | -0.160 | -0.6317 | No | ||
| 34 | PLSCR1 | 1453181_x_at | 19053 | -0.172 | -0.6319 | No | ||
| 35 | ISG20 | 1419569_a_at | 19826 | -0.220 | -0.6512 | No | ||
| 36 | STAT1 | 1420915_at 1440481_at 1450033_a_at 1450034_at | 20572 | -0.307 | -0.6627 | Yes | ||
| 37 | PML | 1448757_at 1456103_at 1459137_at | 21044 | -0.423 | -0.6533 | Yes | ||
| 38 | IRF7 | 1417244_a_at | 21173 | -0.470 | -0.6248 | Yes | ||
| 39 | IFI27 | 1426278_at | 21456 | -0.684 | -0.5877 | Yes | ||
| 40 | SAMHD1 | 1418131_at 1434438_at 1444064_at | 21803 | -1.548 | -0.4905 | Yes | ||
| 41 | TREX1 | 1450672_a_at 1459618_at | 21813 | -1.580 | -0.3755 | Yes | ||
| 42 | PSCD1 | 1418183_a_at 1442189_at 1447786_at 1448997_at 1457095_at | 21931 | -5.215 | 0.0001 | Yes |