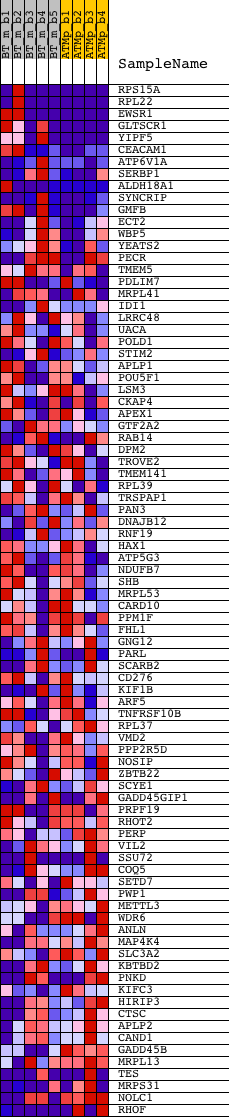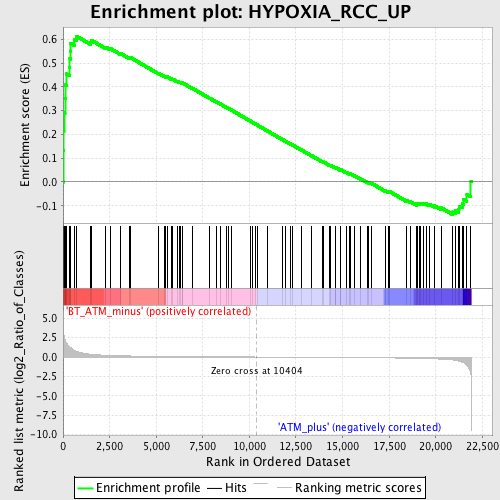
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | HYPOXIA_RCC_UP |
| Enrichment Score (ES) | 0.61316955 |
| Normalized Enrichment Score (NES) | 1.648222 |
| Nominal p-value | 0.0045801527 |
| FDR q-value | 0.4047337 |
| FWER p-Value | 0.997 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RPS15A | 1453467_s_at 1457726_at | 7 | 4.826 | 0.1333 | Yes | ||
| 2 | RPL22 | 1448398_s_at 1453118_s_at | 38 | 2.981 | 0.2145 | Yes | ||
| 3 | EWSR1 | 1417238_at 1425919_at 1436884_x_at 1441074_at 1456461_at | 48 | 2.776 | 0.2910 | Yes | ||
| 4 | GLTSCR1 | 1440238_at 1446548_at 1446616_at 1447189_at | 102 | 2.240 | 0.3506 | Yes | ||
| 5 | YIPF5 | 1418576_at 1444289_at 1449142_a_at | 122 | 2.096 | 0.4078 | Yes | ||
| 6 | CEACAM1 | 1425538_x_at 1425675_s_at 1427630_x_at 1427711_a_at 1427712_at 1450494_x_at 1452532_x_at 1460681_at | 189 | 1.785 | 0.4542 | Yes | ||
| 7 | ATP6V1A | 1422508_at 1445916_at 1450634_at | 357 | 1.302 | 0.4827 | Yes | ||
| 8 | SERBP1 | 1418563_at 1418564_s_at 1418565_at 1437280_s_at 1440195_at 1441501_at | 365 | 1.287 | 0.5180 | Yes | ||
| 9 | ALDH18A1 | 1415836_at 1435855_x_at 1437325_x_at 1437333_x_at 1437620_x_at | 395 | 1.226 | 0.5506 | Yes | ||
| 10 | SYNCRIP | 1422768_at 1422769_at 1426402_at 1428546_at 1450743_s_at | 406 | 1.206 | 0.5835 | Yes | ||
| 11 | GMFB | 1417069_a_at 1431686_a_at 1431687_at 1448570_at 1448571_a_at | 618 | 0.840 | 0.5972 | Yes | ||
| 12 | ECT2 | 1419513_a_at | 712 | 0.732 | 0.6132 | Yes | ||
| 13 | WBP5 | 1451230_a_at | 1487 | 0.355 | 0.5876 | No | ||
| 14 | YEATS2 | 1447943_x_at 1459108_a_at | 1536 | 0.346 | 0.5950 | No | ||
| 15 | PECR | 1439167_at 1448910_at | 2299 | 0.223 | 0.5663 | No | ||
| 16 | TMEM5 | 1424042_at | 2534 | 0.198 | 0.5611 | No | ||
| 17 | PDLIM7 | 1417959_at 1428319_at 1443785_x_at | 3085 | 0.156 | 0.5402 | No | ||
| 18 | MRPL41 | 1428588_a_at 1428589_at 1428590_at | 3583 | 0.132 | 0.5211 | No | ||
| 19 | IDI1 | 1423804_a_at 1451122_at | 3591 | 0.132 | 0.5244 | No | ||
| 20 | LRRC48 | 1424340_at 1440117_at 1457241_at | 5134 | 0.084 | 0.4562 | No | ||
| 21 | UACA | 1452985_at | 5454 | 0.077 | 0.4437 | No | ||
| 22 | POLD1 | 1448187_at 1456055_x_at | 5519 | 0.076 | 0.4429 | No | ||
| 23 | STIM2 | 1434219_at 1444185_at | 5588 | 0.074 | 0.4418 | No | ||
| 24 | APLP1 | 1416134_at 1435857_s_at | 5793 | 0.070 | 0.4344 | No | ||
| 25 | POU5F1 | 1417945_at | 5893 | 0.068 | 0.4318 | No | ||
| 26 | LSM3 | 1448536_at | 6145 | 0.063 | 0.4220 | No | ||
| 27 | CKAP4 | 1426754_x_at 1426755_at 1452181_at 1455019_x_at | 6240 | 0.062 | 0.4195 | No | ||
| 28 | APEX1 | 1416135_at 1437715_x_at 1456079_x_at | 6289 | 0.060 | 0.4189 | No | ||
| 29 | GTF2A2 | 1433528_at | 6405 | 0.058 | 0.4153 | No | ||
| 30 | RAB14 | 1415686_at 1419243_at 1419244_a_at 1419245_at 1419246_s_at 1439687_at 1441992_at | 6415 | 0.058 | 0.4165 | No | ||
| 31 | DPM2 | 1415675_at | 6969 | 0.049 | 0.3925 | No | ||
| 32 | TROVE2 | 1423433_at 1436533_at 1436534_at 1436535_at | 7875 | 0.035 | 0.3521 | No | ||
| 33 | TMEM141 | 1435258_at 1435259_s_at | 8241 | 0.029 | 0.3362 | No | ||
| 34 | RPL39 | 1423032_at 1450840_a_at | 8428 | 0.027 | 0.3284 | No | ||
| 35 | TRSPAP1 | 1428746_a_at 1428747_at | 8756 | 0.022 | 0.3140 | No | ||
| 36 | PAN3 | 1428768_at 1440149_at 1447895_x_at 1458465_at | 8873 | 0.021 | 0.3093 | No | ||
| 37 | DNAJB12 | 1421032_a_at 1435886_at | 9033 | 0.018 | 0.3025 | No | ||
| 38 | RNF19 | 1417508_at 1417509_at | 10053 | 0.005 | 0.2560 | No | ||
| 39 | HAX1 | 1426187_a_at | 10168 | 0.003 | 0.2509 | No | ||
| 40 | ATP5G3 | 1454661_at | 10315 | 0.001 | 0.2442 | No | ||
| 41 | NDUFB7 | 1416417_a_at 1448331_at | 10464 | -0.001 | 0.2375 | No | ||
| 42 | SHB | 1434153_at | 10994 | -0.008 | 0.2135 | No | ||
| 43 | MRPL53 | 1451178_at | 11796 | -0.018 | 0.1773 | No | ||
| 44 | CARD10 | 1449491_at | 11933 | -0.020 | 0.1716 | No | ||
| 45 | PPM1F | 1430631_at 1454934_at | 12186 | -0.023 | 0.1607 | No | ||
| 46 | FHL1 | 1417872_at 1459003_at | 12320 | -0.025 | 0.1553 | No | ||
| 47 | GNG12 | 1421947_at 1455089_at | 12790 | -0.031 | 0.1347 | No | ||
| 48 | PARL | 1433478_at 1447116_at 1447242_at | 13360 | -0.039 | 0.1098 | No | ||
| 49 | SCARB2 | 1454704_at 1460235_at | 13906 | -0.048 | 0.0862 | No | ||
| 50 | CD276 | 1417599_at | 13981 | -0.049 | 0.0841 | No | ||
| 51 | KIF1B | 1423994_at 1423995_at 1425270_at 1451200_at 1451642_at 1451762_a_at 1455182_at 1458426_at | 14329 | -0.054 | 0.0697 | No | ||
| 52 | ARF5 | 1422807_at | 14355 | -0.055 | 0.0701 | No | ||
| 53 | TNFRSF10B | 1421296_at 1422344_s_at | 14609 | -0.059 | 0.0601 | No | ||
| 54 | RPL37 | 1453729_a_at | 14632 | -0.059 | 0.0608 | No | ||
| 55 | VMD2 | 1428841_at | 14878 | -0.063 | 0.0513 | No | ||
| 56 | PPP2R5D | 1450560_a_at | 14894 | -0.063 | 0.0524 | No | ||
| 57 | NOSIP | 1418965_at 1431428_a_at | 15209 | -0.068 | 0.0399 | No | ||
| 58 | ZBTB22 | 1448705_at | 15362 | -0.071 | 0.0349 | No | ||
| 59 | SCYE1 | 1416486_at | 15413 | -0.072 | 0.0346 | No | ||
| 60 | GADD45GIP1 | 1417619_at 1446783_at 1448776_at | 15636 | -0.076 | 0.0265 | No | ||
| 61 | PRPF19 | 1419839_x_at 1449635_at 1456531_x_at 1460633_at | 15973 | -0.082 | 0.0134 | No | ||
| 62 | RHOT2 | 1426822_at | 16363 | -0.090 | -0.0019 | No | ||
| 63 | PERP | 1416271_at 1427378_at | 16419 | -0.091 | -0.0019 | No | ||
| 64 | VIL2 | 1450850_at 1456725_x_at | 16537 | -0.094 | -0.0046 | No | ||
| 65 | SSU72 | 1425674_a_at 1431090_at 1441282_at 1446225_at | 17326 | -0.113 | -0.0375 | No | ||
| 66 | COQ5 | 1417264_at 1417265_s_at | 17481 | -0.118 | -0.0413 | No | ||
| 67 | SETD7 | 1423003_at 1435437_at | 17536 | -0.119 | -0.0405 | No | ||
| 68 | PWP1 | 1417873_at 1454142_a_at | 18424 | -0.146 | -0.0770 | No | ||
| 69 | METTL3 | 1423099_a_at | 18631 | -0.154 | -0.0822 | No | ||
| 70 | WDR6 | 1415770_at 1455940_x_at | 18970 | -0.168 | -0.0930 | No | ||
| 71 | ANLN | 1433543_at 1439648_at | 19014 | -0.170 | -0.0903 | No | ||
| 72 | MAP4K4 | 1422615_at 1440609_at 1448050_s_at 1459912_at | 19132 | -0.176 | -0.0908 | No | ||
| 73 | SLC3A2 | 1425364_a_at | 19196 | -0.179 | -0.0887 | No | ||
| 74 | KBTBD2 | 1451214_at | 19370 | -0.188 | -0.0914 | No | ||
| 75 | PNKD | 1418746_at 1451399_at | 19491 | -0.196 | -0.0915 | No | ||
| 76 | KIFC3 | 1416199_at | 19668 | -0.209 | -0.0937 | No | ||
| 77 | HIRIP3 | 1434936_at | 19927 | -0.230 | -0.0992 | No | ||
| 78 | CTSC | 1416382_at 1437939_s_at 1446834_at | 20312 | -0.271 | -0.1092 | No | ||
| 79 | APLP2 | 1421887_a_at 1421888_x_at 1421889_a_at 1423739_x_at 1432344_a_at 1446786_at | 20914 | -0.379 | -0.1262 | No | ||
| 80 | CAND1 | 1428648_at 1428649_at 1435781_at 1455751_at | 21062 | -0.428 | -0.1211 | No | ||
| 81 | GADD45B | 1420197_at 1449773_s_at 1450971_at | 21244 | -0.512 | -0.1152 | No | ||
| 82 | MRPL13 | 1424204_at 1435966_x_at 1459856_at 1460354_a_at | 21260 | -0.522 | -0.1015 | No | ||
| 83 | TES | 1424246_a_at 1440649_at 1460378_a_at | 21431 | -0.662 | -0.0909 | No | ||
| 84 | MRPS31 | 1417737_at 1443165_at | 21492 | -0.727 | -0.0735 | No | ||
| 85 | NOLC1 | 1428869_at 1428870_at 1450087_a_at | 21686 | -1.076 | -0.0525 | No | ||
| 86 | RHOF | 1434794_at 1441510_at 1455902_x_at | 21901 | -2.305 | 0.0015 | No |