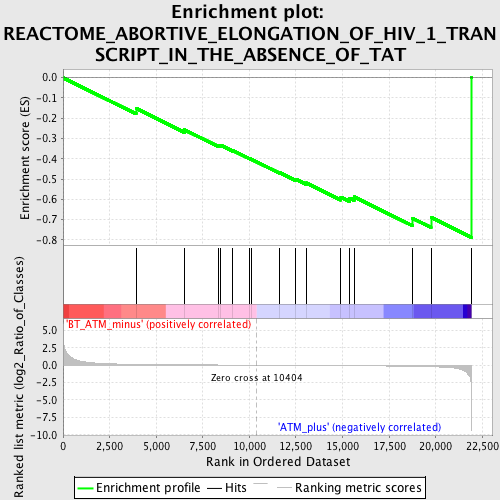
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus.phenotype_BT_ATM_minus_versus_ATM_plus.cls #BT_ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_BT_ATM_minus_versus_ATM_plus.cls#BT_ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | REACTOME_ABORTIVE_ELONGATION_OF_HIV_1_TRANSCRIPT_IN_THE_ABSENCE_OF_TAT |
| Enrichment Score (ES) | -0.7876132 |
| Normalized Enrichment Score (NES) | -1.7586635 |
| Nominal p-value | 0.0045351475 |
| FDR q-value | 0.19745839 |
| FWER p-Value | 0.86 |

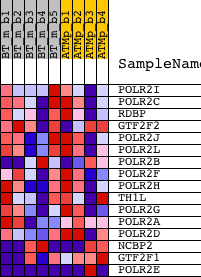
| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | POLR2I | 1428494_a_at | 3920 | 0.119 | -0.1515 | No | ||
| 2 | POLR2C | 1416341_at | 6514 | 0.057 | -0.2567 | No | ||
| 3 | RDBP | 1450705_at | 8370 | 0.027 | -0.3350 | No | ||
| 4 | GTF2F2 | 1426626_at 1443233_at | 8469 | 0.026 | -0.3334 | No | ||
| 5 | POLR2J | 1417720_at 1440065_at | 9098 | 0.018 | -0.3580 | No | ||
| 6 | POLR2L | 1428296_at | 10020 | 0.005 | -0.3988 | No | ||
| 7 | POLR2B | 1433552_a_at | 10116 | 0.004 | -0.4023 | No | ||
| 8 | POLR2F | 1415754_at 1427402_at | 11639 | -0.016 | -0.4680 | No | ||
| 9 | POLR2H | 1424473_at | 12464 | -0.027 | -0.4995 | No | ||
| 10 | TH1L | 1416198_at | 13068 | -0.035 | -0.5189 | No | ||
| 11 | POLR2G | 1416270_at | 14915 | -0.064 | -0.5884 | No | ||
| 12 | POLR2A | 1422311_a_at 1426242_at 1458710_at | 15394 | -0.072 | -0.5937 | No | ||
| 13 | POLR2D | 1424258_at | 15625 | -0.075 | -0.5868 | No | ||
| 14 | NCBP2 | 1423045_at 1423046_s_at 1450847_at | 18754 | -0.159 | -0.6927 | Yes | ||
| 15 | GTF2F1 | 1417698_at 1417699_at | 19758 | -0.214 | -0.6889 | Yes | ||
| 16 | POLR2E | 1417138_s_at 1451093_at 1458326_at | 21922 | -3.411 | 0.0005 | Yes |