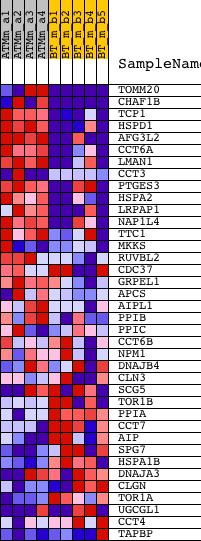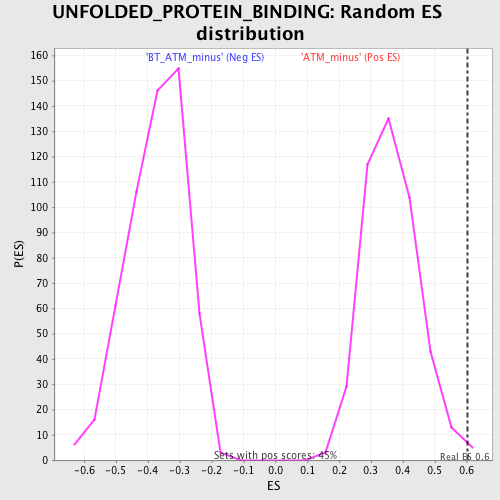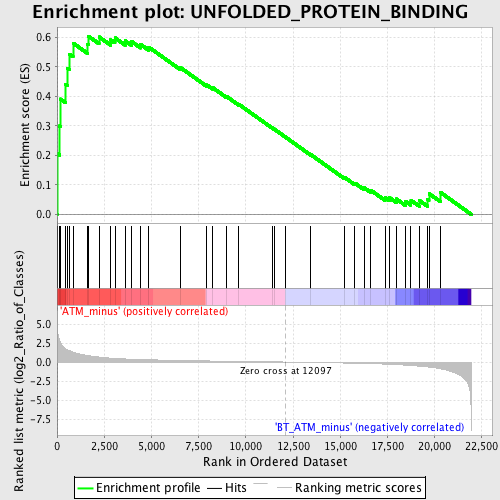
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | UNFOLDED_PROTEIN_BINDING |
| Enrichment Score (ES) | 0.6039833 |
| Normalized Enrichment Score (NES) | 1.6584069 |
| Nominal p-value | 0.008908686 |
| FDR q-value | 0.30776432 |
| FWER p-Value | 0.985 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TOMM20 | 1423080_at 1423081_a_at 1435187_at 1435534_a_at 1455357_x_at 1458692_at | 6 | 5.785 | 0.2065 | Yes | ||
| 2 | CHAF1B | 1423877_at 1431275_at | 134 | 2.769 | 0.2997 | Yes | ||
| 3 | TCP1 | 1442317_at 1448122_at | 166 | 2.596 | 0.3910 | Yes | ||
| 4 | HSPD1 | 1426351_at | 433 | 1.757 | 0.4417 | Yes | ||
| 5 | AFG3L2 | 1427206_at 1427207_s_at 1454003_at | 535 | 1.626 | 0.4952 | Yes | ||
| 6 | CCT6A | 1423517_at 1455988_a_at | 667 | 1.476 | 0.5420 | Yes | ||
| 7 | LMAN1 | 1428129_at 1428130_at 1444037_at 1452671_s_at | 845 | 1.300 | 0.5803 | Yes | ||
| 8 | CCT3 | 1416024_x_at 1426067_x_at 1439679_at 1448178_a_at 1449645_s_at 1451915_at 1459987_s_at | 1614 | 0.850 | 0.5756 | Yes | ||
| 9 | PTGES3 | 1417998_at 1460221_at | 1651 | 0.839 | 0.6040 | Yes | ||
| 10 | HSPA2 | 1417101_at | 2222 | 0.650 | 0.6012 | No | ||
| 11 | LRPAP1 | 1426696_at 1426697_a_at 1436609_a_at 1452148_at | 2840 | 0.511 | 0.5913 | No | ||
| 12 | NAP1L4 | 1448476_at | 3066 | 0.474 | 0.5980 | No | ||
| 13 | TTC1 | 1416994_at 1448537_at | 3602 | 0.403 | 0.5879 | No | ||
| 14 | MKKS | 1422627_a_at 1454014_a_at | 3932 | 0.370 | 0.5861 | No | ||
| 15 | RUVBL2 | 1422482_at | 4425 | 0.330 | 0.5755 | No | ||
| 16 | CDC37 | 1416819_at 1416820_at | 4854 | 0.300 | 0.5666 | No | ||
| 17 | GRPEL1 | 1417320_at | 6520 | 0.204 | 0.4979 | No | ||
| 18 | APCS | 1419059_at | 7928 | 0.143 | 0.4387 | No | ||
| 19 | AIPL1 | 1425590_s_at 1427952_at | 8240 | 0.132 | 0.4292 | No | ||
| 20 | PPIB | 1437649_x_at 1450911_at | 8957 | 0.106 | 0.4003 | No | ||
| 21 | PPIC | 1416498_at | 9631 | 0.083 | 0.3725 | No | ||
| 22 | CCT6B | 1419227_at | 11388 | 0.024 | 0.2932 | No | ||
| 23 | NPM1 | 1415839_a_at 1420267_at 1420268_x_at 1420269_at 1432416_a_at | 11489 | 0.021 | 0.2894 | No | ||
| 24 | DNAJB4 | 1431734_a_at 1439524_at 1443611_at 1451177_at | 12112 | -0.001 | 0.2610 | No | ||
| 25 | CLN3 | 1417551_at 1457789_at | 13438 | -0.050 | 0.2023 | No | ||
| 26 | SCG5 | 1423150_at 1446755_at | 15203 | -0.127 | 0.1263 | No | ||
| 27 | TOR1B | 1417819_at 1417820_at 1448848_at 1457114_at | 15763 | -0.156 | 0.1063 | No | ||
| 28 | PPIA | 1417451_a_at | 16259 | -0.186 | 0.0903 | No | ||
| 29 | CCT7 | 1415816_at 1415817_s_at | 16608 | -0.210 | 0.0820 | No | ||
| 30 | AIP | 1423128_at | 17367 | -0.270 | 0.0570 | No | ||
| 31 | SPG7 | 1441585_at | 17613 | -0.295 | 0.0564 | No | ||
| 32 | HSPA1B | 1427126_at 1427127_x_at 1452318_a_at | 17962 | -0.336 | 0.0525 | No | ||
| 33 | DNAJA3 | 1420629_a_at 1432066_at 1449935_a_at | 18456 | -0.407 | 0.0445 | No | ||
| 34 | CLGN | 1418617_x_at 1430240_a_at 1457829_at | 18739 | -0.455 | 0.0479 | No | ||
| 35 | TOR1A | 1426515_a_at | 19183 | -0.546 | 0.0472 | No | ||
| 36 | UGCGL1 | 1432318_at 1448009_at 1455839_at | 19612 | -0.643 | 0.0506 | No | ||
| 37 | CCT4 | 1415867_at 1415868_at 1430034_at 1433447_x_at 1438560_x_at 1442569_at 1456082_x_at 1456572_x_at | 19696 | -0.664 | 0.0706 | No | ||
| 38 | TAPBP | 1421812_at 1450378_at | 20305 | -0.884 | 0.0744 | No |