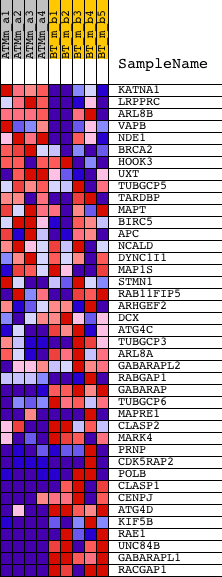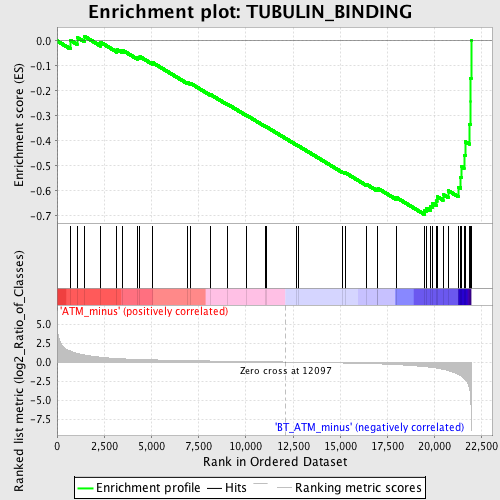
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | TUBULIN_BINDING |
| Enrichment Score (ES) | -0.69415593 |
| Normalized Enrichment Score (NES) | -1.9110821 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.07501495 |
| FWER p-Value | 0.072 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | KATNA1 | 1450949_at | 699 | 1.434 | 0.0026 | No | ||
| 2 | LRPPRC | 1424353_at | 1059 | 1.142 | 0.0138 | No | ||
| 3 | ARL8B | 1418819_at 1428137_at 1428138_s_at | 1432 | 0.927 | 0.0191 | No | ||
| 4 | VAPB | 1423152_at 1436079_s_at 1445370_at 1458501_at | 2305 | 0.627 | -0.0056 | No | ||
| 5 | NDE1 | 1435737_a_at | 3170 | 0.460 | -0.0340 | No | ||
| 6 | BRCA2 | 1419076_a_at | 3462 | 0.420 | -0.0371 | No | ||
| 7 | HOOK3 | 1443857_at 1446737_a_at | 4255 | 0.342 | -0.0650 | No | ||
| 8 | UXT | 1418986_a_at | 4387 | 0.333 | -0.0630 | No | ||
| 9 | TUBGCP5 | 1426518_at 1439923_at 1439924_x_at 1457282_x_at | 5077 | 0.287 | -0.0876 | No | ||
| 10 | TARDBP | 1423723_s_at 1428467_at 1434419_s_at 1436318_at 1455655_a_at | 6884 | 0.186 | -0.1656 | No | ||
| 11 | MAPT | 1417885_at 1424718_at 1424719_a_at 1445634_at 1455028_at | 7076 | 0.179 | -0.1700 | No | ||
| 12 | BIRC5 | 1424278_a_at | 8131 | 0.135 | -0.2148 | No | ||
| 13 | APC | 1420956_at 1420957_at 1435543_at 1450056_at | 9040 | 0.103 | -0.2538 | No | ||
| 14 | NCALD | 1417568_at 1417569_at 1439730_at 1444466_at 1458037_at | 10034 | 0.069 | -0.2975 | No | ||
| 15 | DYNC1I1 | 1416361_a_at | 11012 | 0.036 | -0.3413 | No | ||
| 16 | MAP1S | 1454732_at | 11090 | 0.034 | -0.3440 | No | ||
| 17 | STMN1 | 1415849_s_at | 12666 | -0.021 | -0.4154 | No | ||
| 18 | RAB11FIP5 | 1427405_s_at 1434314_s_at | 12789 | -0.026 | -0.4203 | No | ||
| 19 | ARHGEF2 | 1421042_at 1421043_s_at 1427646_a_at | 15138 | -0.123 | -0.5246 | No | ||
| 20 | DCX | 1418139_at 1418140_at 1418141_at 1448974_at | 15267 | -0.130 | -0.5273 | No | ||
| 21 | ATG4C | 1434014_at 1441440_at | 16401 | -0.196 | -0.5744 | No | ||
| 22 | TUBGCP3 | 1426707_at | 16961 | -0.236 | -0.5942 | No | ||
| 23 | ARL8A | 1424640_at | 16992 | -0.239 | -0.5898 | No | ||
| 24 | GABARAPL2 | 1423187_at 1441376_at 1444956_at 1459477_at | 17957 | -0.335 | -0.6258 | No | ||
| 25 | RABGAP1 | 1424188_at 1441217_at 1441800_at 1443535_at 1457370_at 1460486_at | 19455 | -0.601 | -0.6797 | Yes | ||
| 26 | GABARAP | 1416937_at | 19548 | -0.625 | -0.6688 | Yes | ||
| 27 | TUBGCP6 | 1435090_at | 19778 | -0.689 | -0.6626 | Yes | ||
| 28 | MAPRE1 | 1422764_at 1422765_at 1428819_at 1428820_at 1450740_a_at | 19895 | -0.725 | -0.6504 | Yes | ||
| 29 | CLASP2 | 1424836_a_at 1427328_a_at 1429976_at 1440802_at 1441726_at 1443320_at 1456911_at | 20084 | -0.792 | -0.6399 | Yes | ||
| 30 | MARK4 | 1439051_a_at 1456618_at | 20120 | -0.803 | -0.6222 | Yes | ||
| 31 | PRNP | 1416130_at 1446507_at 1448233_at | 20440 | -0.945 | -0.6140 | Yes | ||
| 32 | CDK5RAP2 | 1443976_at 1447103_at 1452618_at | 20728 | -1.144 | -0.5995 | Yes | ||
| 33 | POLB | 1425371_at 1434229_a_at | 21264 | -1.646 | -0.5843 | Yes | ||
| 34 | CLASP1 | 1427353_at 1452265_at 1459233_at 1460102_at | 21370 | -1.793 | -0.5458 | Yes | ||
| 35 | CENPJ | 1423140_at 1423141_at 1437643_at 1450872_s_at | 21410 | -1.876 | -0.5024 | Yes | ||
| 36 | ATG4D | 1433918_at 1439130_at | 21552 | -2.170 | -0.4565 | Yes | ||
| 37 | KIF5B | 1418427_at 1418428_at 1418429_at 1418430_at 1418431_at | 21625 | -2.334 | -0.4035 | Yes | ||
| 38 | RAE1 | 1429528_at 1444256_at 1459822_at | 21824 | -3.273 | -0.3337 | Yes | ||
| 39 | UNC84B | 1433832_at 1439238_at | 21879 | -3.868 | -0.2429 | Yes | ||
| 40 | GABARAPL1 | 1416418_at 1416419_s_at 1442572_at | 21881 | -3.884 | -0.1493 | Yes | ||
| 41 | RACGAP1 | 1421546_a_at 1441889_x_at 1451358_a_at | 21926 | -6.293 | 0.0004 | Yes |