Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | ATM_minus |
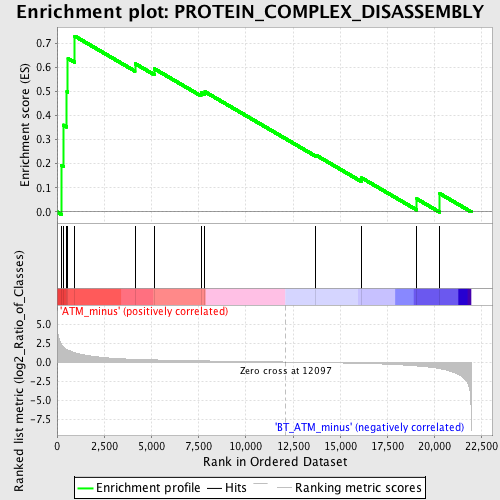
| GeneSet | PROTEIN_COMPLEX_DISASSEMBLY |
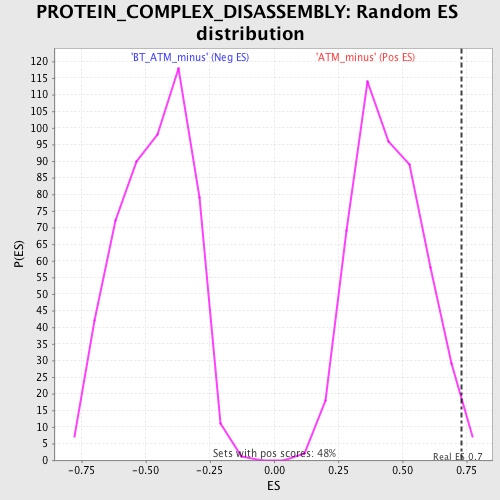
| Enrichment Score (ES) | 0.7289691 |
| Normalized Enrichment Score (NES) | 1.6374793 |
| Nominal p-value | 0.014522822 |
| FDR q-value | 0.33458188 |
| FWER p-Value | 0.994 |

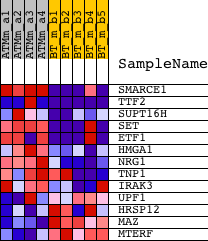
| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SMARCE1 | 1422675_at 1422676_at 1430822_at | 232 | 2.271 | 0.1920 | Yes | ||
| 2 | TTF2 | 1428522_at 1447119_at 1452818_at | 331 | 1.939 | 0.3605 | Yes | ||
| 3 | SUPT16H | 1419741_at 1449578_at 1456449_at | 520 | 1.651 | 0.4992 | Yes | ||
| 4 | SET | 1426853_at 1426854_a_at | 575 | 1.567 | 0.6365 | Yes | ||
| 5 | ETF1 | 1420023_at 1420024_s_at 1424013_at 1440629_at 1451208_at | 938 | 1.222 | 0.7290 | Yes | ||
| 6 | HMGA1 | 1416184_s_at | 4125 | 0.354 | 0.6152 | No | ||
| 7 | NRG1 | 1456524_at | 5146 | 0.282 | 0.5938 | No | ||
| 8 | TNP1 | 1415924_at 1438632_x_at | 7623 | 0.154 | 0.4946 | No | ||
| 9 | IRAK3 | 1430704_at 1435040_at | 7830 | 0.147 | 0.4983 | No | ||
| 10 | UPF1 | 1419685_at | 13702 | -0.060 | 0.2359 | No | ||
| 11 | HRSP12 | 1419348_at 1428326_s_at | 16131 | -0.179 | 0.1410 | No | ||
| 12 | MAZ | 1427099_at | 19052 | -0.517 | 0.0540 | No | ||
| 13 | MTERF | 1429698_at | 20269 | -0.868 | 0.0760 | No |