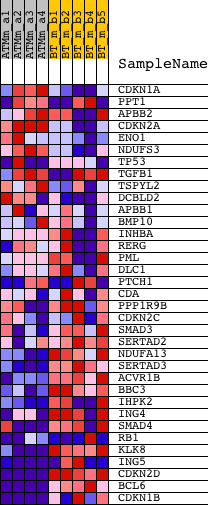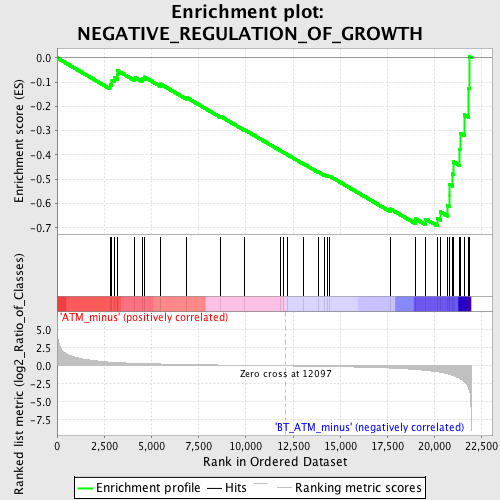
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | NEGATIVE_REGULATION_OF_GROWTH |
| Enrichment Score (ES) | -0.6927798 |
| Normalized Enrichment Score (NES) | -1.8257093 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.18943112 |
| FWER p-Value | 0.311 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CDKN1A | 1421679_a_at 1424638_at | 2802 | 0.519 | -0.1079 | No | ||
| 2 | PPT1 | 1420015_s_at 1420016_at 1422467_at 1422468_at 1444884_at | 2903 | 0.502 | -0.0930 | No | ||
| 3 | APBB2 | 1426719_at 1426720_at 1440135_at 1446481_at 1452342_at | 3035 | 0.479 | -0.0804 | No | ||
| 4 | CDKN2A | 1450140_a_at | 3186 | 0.458 | -0.0696 | No | ||
| 5 | ENO1 | 1419023_x_at | 3198 | 0.457 | -0.0524 | No | ||
| 6 | NDUFS3 | 1423737_at | 4123 | 0.354 | -0.0809 | No | ||
| 7 | TP53 | 1426538_a_at 1427739_a_at 1438808_at 1457623_x_at 1459780_at 1459781_x_at | 4498 | 0.325 | -0.0854 | No | ||
| 8 | TGFB1 | 1420653_at 1445360_at | 4620 | 0.315 | -0.0787 | No | ||
| 9 | TSPYL2 | 1434849_at | 5472 | 0.261 | -0.1075 | No | ||
| 10 | DCBLD2 | 1420526_at 1437635_at 1449891_a_at | 6864 | 0.187 | -0.1638 | No | ||
| 11 | APBB1 | 1423892_at 1423893_x_at | 8666 | 0.116 | -0.2415 | No | ||
| 12 | BMP10 | 1421763_at | 9899 | 0.074 | -0.2949 | No | ||
| 13 | INHBA | 1422053_at 1458291_at | 11826 | 0.009 | -0.3825 | No | ||
| 14 | RERG | 1447446_at 1451236_at | 12003 | 0.003 | -0.3904 | No | ||
| 15 | PML | 1448757_at 1456103_at 1459137_at | 12182 | -0.003 | -0.3984 | No | ||
| 16 | DLC1 | 1436173_at 1450206_at 1458220_at 1460602_at | 13066 | -0.036 | -0.4373 | No | ||
| 17 | PTCH1 | 1428853_at 1439663_at 1447039_at 1450824_at | 13817 | -0.065 | -0.4690 | No | ||
| 18 | CDA | 1427357_at | 14153 | -0.079 | -0.4813 | No | ||
| 19 | PPP1R9B | 1447247_at 1447486_at | 14322 | -0.086 | -0.4856 | No | ||
| 20 | CDKN2C | 1416868_at 1439164_at | 14434 | -0.091 | -0.4871 | No | ||
| 21 | SMAD3 | 1450471_at 1450472_s_at 1454960_at | 17658 | -0.300 | -0.6227 | No | ||
| 22 | SERTAD2 | 1417209_at 1444645_at 1454815_at | 18966 | -0.502 | -0.6630 | No | ||
| 23 | NDUFA13 | 1430713_s_at | 19515 | -0.615 | -0.6642 | Yes | ||
| 24 | SERTAD3 | 1421076_at 1421077_at | 20142 | -0.811 | -0.6614 | Yes | ||
| 25 | ACVR1B | 1422098_at 1433725_at | 20303 | -0.883 | -0.6345 | Yes | ||
| 26 | BBC3 | 1423315_at | 20687 | -1.114 | -0.6089 | Yes | ||
| 27 | IHPK2 | 1428373_at 1435319_at | 20792 | -1.186 | -0.5677 | Yes | ||
| 28 | ING4 | 1448054_at 1460728_s_at | 20793 | -1.187 | -0.5218 | Yes | ||
| 29 | SMAD4 | 1422485_at 1422486_a_at 1422487_at 1444205_at | 20941 | -1.295 | -0.4784 | Yes | ||
| 30 | RB1 | 1417850_at 1441494_at 1444400_at 1444454_at 1457447_at | 20989 | -1.340 | -0.4287 | Yes | ||
| 31 | KLK8 | 1419722_at | 21332 | -1.735 | -0.3772 | Yes | ||
| 32 | ING5 | 1431134_at 1436069_at 1442094_at 1454235_a_at | 21338 | -1.743 | -0.3099 | Yes | ||
| 33 | CDKN2D | 1416253_at | 21575 | -2.228 | -0.2345 | Yes | ||
| 34 | BCL6 | 1421818_at 1450381_a_at | 21792 | -3.059 | -0.1260 | Yes | ||
| 35 | CDKN1B | 1419497_at 1434045_at | 21841 | -3.422 | 0.0042 | Yes |