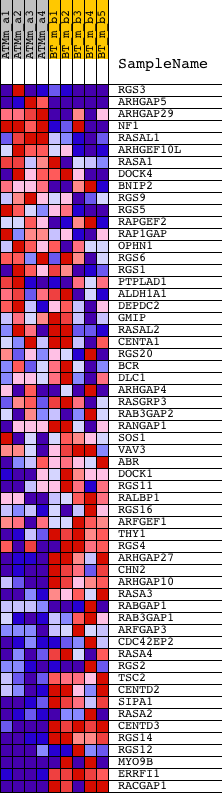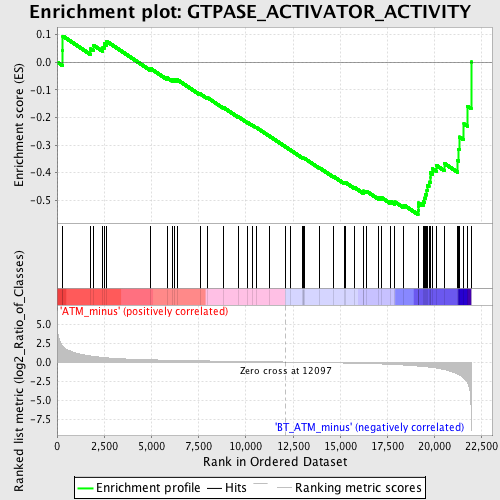
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | GTPASE_ACTIVATOR_ACTIVITY |
| Enrichment Score (ES) | -0.55108374 |
| Normalized Enrichment Score (NES) | -1.6250695 |
| Nominal p-value | 0.0018621974 |
| FDR q-value | 0.32658586 |
| FWER p-Value | 0.998 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RGS3 | 1425701_a_at 1427648_at 1449516_a_at 1454026_a_at | 297 | 2.018 | 0.0406 | No | ||
| 2 | ARHGAP5 | 1423194_at 1450896_at 1450897_at 1457410_at | 306 | 1.997 | 0.0938 | No | ||
| 3 | ARHGAP29 | 1441618_at 1444512_at 1454745_at | 1781 | 0.785 | 0.0475 | No | ||
| 4 | NF1 | 1438067_at 1441523_at 1443097_at 1452525_a_at | 1915 | 0.738 | 0.0612 | No | ||
| 5 | RASAL1 | 1417858_at | 2425 | 0.598 | 0.0540 | No | ||
| 6 | ARHGEF10L | 1460405_at | 2502 | 0.578 | 0.0661 | No | ||
| 7 | RASA1 | 1426476_at 1426477_at 1426478_at 1438998_at | 2630 | 0.549 | 0.0750 | No | ||
| 8 | DOCK4 | 1431114_at 1436405_at 1441462_at 1441469_at 1446283_at 1457146_at 1459279_at | 4970 | 0.293 | -0.0241 | No | ||
| 9 | BNIP2 | 1422490_at 1422491_a_at 1431731_at 1433025_x_at 1441989_at 1453993_a_at 1454441_at 1454442_at | 5822 | 0.239 | -0.0565 | No | ||
| 10 | RGS9 | 1418691_at 1426033_at 1439635_at | 6090 | 0.225 | -0.0627 | No | ||
| 11 | RGS5 | 1420940_x_at 1420941_at | 6226 | 0.218 | -0.0630 | No | ||
| 12 | RAPGEF2 | 1452833_at | 6366 | 0.212 | -0.0637 | No | ||
| 13 | RAP1GAP | 1428443_a_at | 7569 | 0.156 | -0.1145 | No | ||
| 14 | OPHN1 | 1419107_at 1419108_at 1456709_at | 7964 | 0.142 | -0.1287 | No | ||
| 15 | RGS6 | 1427340_at 1452399_at | 8837 | 0.110 | -0.1656 | No | ||
| 16 | RGS1 | 1417601_at | 9598 | 0.084 | -0.1981 | No | ||
| 17 | PTPLAD1 | 1433706_a_at 1452427_s_at 1454698_at | 10091 | 0.068 | -0.2187 | No | ||
| 18 | ALDH1A1 | 1416468_at 1445980_at | 10332 | 0.060 | -0.2281 | No | ||
| 19 | DEPDC2 | 1432047_at | 10533 | 0.052 | -0.2358 | No | ||
| 20 | GMIP | 1428784_at | 11232 | 0.029 | -0.2670 | No | ||
| 21 | RASAL2 | 1436910_at 1444671_at | 12070 | 0.001 | -0.3052 | No | ||
| 22 | CENTA1 | 1433556_at | 12352 | -0.010 | -0.3178 | No | ||
| 23 | RGS20 | 1421493_a_at 1443694_at 1446199_at | 12969 | -0.033 | -0.3451 | No | ||
| 24 | BCR | 1427265_at 1452368_at 1455564_at | 13026 | -0.035 | -0.3467 | No | ||
| 25 | DLC1 | 1436173_at 1450206_at 1458220_at 1460602_at | 13066 | -0.036 | -0.3475 | No | ||
| 26 | ARHGAP4 | 1419296_at 1443360_x_at 1458190_at | 13091 | -0.037 | -0.3476 | No | ||
| 27 | RASGRP3 | 1438030_at 1438031_at | 13903 | -0.068 | -0.3828 | No | ||
| 28 | RAB3GAP2 | 1436272_at 1454915_at | 14627 | -0.101 | -0.4132 | No | ||
| 29 | RANGAP1 | 1423749_s_at 1444581_at 1451092_a_at | 15201 | -0.127 | -0.4360 | No | ||
| 30 | SOS1 | 1421884_at 1421885_at 1421886_at | 15287 | -0.131 | -0.4364 | No | ||
| 31 | VAV3 | 1417122_at 1417123_at 1446795_at 1448600_s_at 1458630_at | 15772 | -0.157 | -0.4543 | No | ||
| 32 | ABR | 1433477_at | 16212 | -0.183 | -0.4694 | No | ||
| 33 | DOCK1 | 1443390_at 1443760_at 1443991_at 1452220_at 1457806_at | 16249 | -0.185 | -0.4661 | No | ||
| 34 | RGS11 | 1425245_a_at | 16385 | -0.194 | -0.4671 | No | ||
| 35 | RALBP1 | 1417248_at 1448634_at | 17024 | -0.241 | -0.4898 | No | ||
| 36 | RGS16 | 1426037_a_at 1451452_a_at 1455265_a_at | 17165 | -0.253 | -0.4894 | No | ||
| 37 | ARFGEF1 | 1415711_at 1437104_at 1444175_at 1456917_at | 17633 | -0.297 | -0.5027 | No | ||
| 38 | THY1 | 1423135_at | 17892 | -0.326 | -0.5058 | No | ||
| 39 | RGS4 | 1416286_at 1416287_at 1448285_at | 18366 | -0.392 | -0.5169 | No | ||
| 40 | ARHGAP27 | 1416758_at 1457655_x_at | 19115 | -0.532 | -0.5368 | Yes | ||
| 41 | CHN2 | 1428573_at 1428574_a_at 1441158_at 1443626_at | 19141 | -0.537 | -0.5235 | Yes | ||
| 42 | ARHGAP10 | 1426027_a_at 1440347_at 1459609_at | 19148 | -0.538 | -0.5094 | Yes | ||
| 43 | RASA3 | 1415850_at | 19387 | -0.587 | -0.5045 | Yes | ||
| 44 | RABGAP1 | 1424188_at 1441217_at 1441800_at 1443535_at 1457370_at 1460486_at | 19455 | -0.601 | -0.4914 | Yes | ||
| 45 | RAB3GAP1 | 1455922_at | 19519 | -0.616 | -0.4778 | Yes | ||
| 46 | ARFGAP3 | 1426534_a_at 1430904_at | 19585 | -0.637 | -0.4637 | Yes | ||
| 47 | CDC42EP2 | 1428750_at 1445870_at | 19611 | -0.643 | -0.4475 | Yes | ||
| 48 | RASA4 | 1417333_at 1441458_at | 19698 | -0.665 | -0.4336 | Yes | ||
| 49 | RGS2 | 1419247_at 1419248_at 1447830_s_at | 19766 | -0.686 | -0.4183 | Yes | ||
| 50 | TSC2 | 1452105_a_at | 19769 | -0.686 | -0.3999 | Yes | ||
| 51 | CENTD2 | 1428064_at | 19891 | -0.723 | -0.3861 | Yes | ||
| 52 | SIPA1 | 1416206_at | 20083 | -0.792 | -0.3736 | Yes | ||
| 53 | RASA2 | 1422785_at 1455181_at 1457586_at | 20518 | -0.985 | -0.3669 | Yes | ||
| 54 | CENTD3 | 1419833_s_at 1447407_at 1451282_at | 21202 | -1.573 | -0.3560 | Yes | ||
| 55 | RGS14 | 1419221_a_at | 21271 | -1.651 | -0.3148 | Yes | ||
| 56 | RGS12 | 1429380_at 1453129_a_at 1457511_at 1459566_at | 21311 | -1.703 | -0.2708 | Yes | ||
| 57 | MYO9B | 1418031_at 1438533_at 1459110_at | 21545 | -2.156 | -0.2236 | Yes | ||
| 58 | ERRFI1 | 1416129_at 1419816_s_at | 21725 | -2.698 | -0.1594 | Yes | ||
| 59 | RACGAP1 | 1421546_a_at 1441889_x_at 1451358_a_at | 21926 | -6.293 | 0.0004 | Yes |