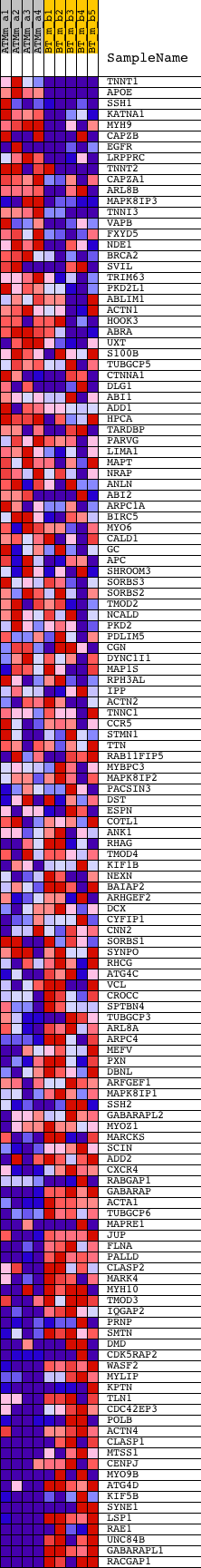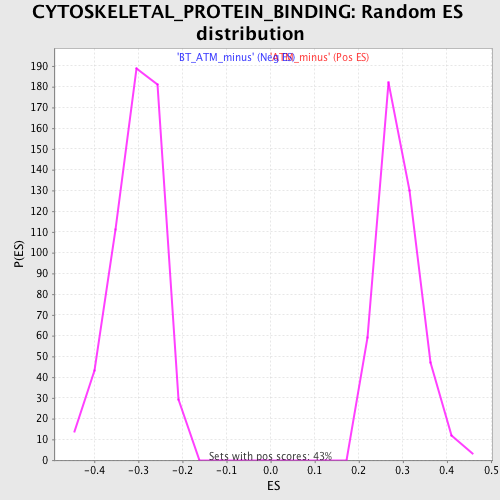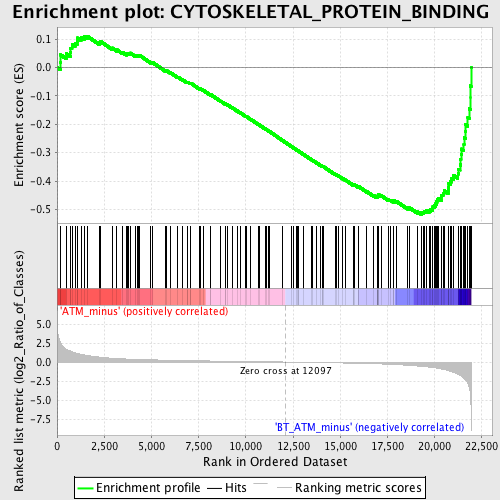
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | CYTOSKELETAL_PROTEIN_BINDING |
| Enrichment Score (ES) | -0.51790524 |
| Normalized Enrichment Score (NES) | -1.711545 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.27328268 |
| FWER p-Value | 0.884 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TNNT1 | 1419606_a_at | 182 | 2.528 | 0.0186 | No | ||
| 2 | APOE | 1432466_a_at | 191 | 2.473 | 0.0446 | No | ||
| 3 | SSH1 | 1433874_at 1438253_at 1439237_a_at 1455854_a_at | 484 | 1.694 | 0.0492 | No | ||
| 4 | KATNA1 | 1450949_at | 699 | 1.434 | 0.0547 | No | ||
| 5 | MYH9 | 1417472_at 1420170_at 1420171_s_at 1420172_at 1440708_at | 722 | 1.411 | 0.0687 | No | ||
| 6 | CAPZB | 1417259_a_at 1424168_a_at 1431668_at 1453960_a_at | 789 | 1.354 | 0.0801 | No | ||
| 7 | EGFR | 1424932_at 1432647_at 1435888_at 1451530_at 1454313_at 1457563_at 1460420_a_at | 951 | 1.215 | 0.0856 | No | ||
| 8 | LRPPRC | 1424353_at | 1059 | 1.142 | 0.0929 | No | ||
| 9 | TNNT2 | 1418726_a_at 1424967_x_at 1440424_at | 1076 | 1.129 | 0.1042 | No | ||
| 10 | CAPZA1 | 1439455_x_at 1440380_at | 1267 | 1.016 | 0.1063 | No | ||
| 11 | ARL8B | 1418819_at 1428137_at 1428138_s_at | 1432 | 0.927 | 0.1086 | No | ||
| 12 | MAPK8IP3 | 1416437_a_at 1425975_a_at 1436676_at 1460628_at | 1588 | 0.858 | 0.1107 | No | ||
| 13 | TNNI3 | 1422536_at | 2241 | 0.643 | 0.0876 | No | ||
| 14 | VAPB | 1423152_at 1436079_s_at 1445370_at 1458501_at | 2305 | 0.627 | 0.0914 | No | ||
| 15 | FXYD5 | 1418296_at | 2913 | 0.500 | 0.0689 | No | ||
| 16 | NDE1 | 1435737_a_at | 3170 | 0.460 | 0.0620 | No | ||
| 17 | BRCA2 | 1419076_a_at | 3462 | 0.420 | 0.0532 | No | ||
| 18 | SVIL | 1427662_at 1460694_s_at 1460735_at | 3666 | 0.396 | 0.0481 | No | ||
| 19 | TRIM63 | 1438704_at | 3705 | 0.392 | 0.0505 | No | ||
| 20 | PKD2L1 | 1457105_at | 3783 | 0.384 | 0.0511 | No | ||
| 21 | ABLIM1 | 1442376_at 1453103_at 1454708_at 1460120_at | 3868 | 0.375 | 0.0512 | No | ||
| 22 | ACTN1 | 1427385_s_at 1428585_at 1452415_at | 4153 | 0.351 | 0.0419 | No | ||
| 23 | HOOK3 | 1443857_at 1446737_a_at | 4255 | 0.342 | 0.0409 | No | ||
| 24 | ABRA | 1458455_at | 4322 | 0.336 | 0.0415 | No | ||
| 25 | UXT | 1418986_a_at | 4387 | 0.333 | 0.0421 | No | ||
| 26 | S100B | 1419383_at 1434342_at | 4969 | 0.293 | 0.0185 | No | ||
| 27 | TUBGCP5 | 1426518_at 1439923_at 1439924_x_at 1457282_x_at | 5077 | 0.287 | 0.0167 | No | ||
| 28 | CTNNA1 | 1437275_at 1437807_x_at 1443662_at 1448149_at | 5748 | 0.243 | -0.0115 | No | ||
| 29 | DLG1 | 1415691_at 1445798_at 1450768_at 1459635_at | 5794 | 0.241 | -0.0110 | No | ||
| 30 | ABI1 | 1423177_a_at 1423178_at 1450890_a_at | 5999 | 0.230 | -0.0179 | No | ||
| 31 | ADD1 | 1420953_at 1420954_a_at 1439125_at 1450054_at | 6395 | 0.210 | -0.0338 | No | ||
| 32 | HPCA | 1425833_a_at 1450930_at | 6643 | 0.198 | -0.0430 | No | ||
| 33 | TARDBP | 1423723_s_at 1428467_at 1434419_s_at 1436318_at 1455655_a_at | 6884 | 0.186 | -0.0520 | No | ||
| 34 | PARVG | 1416875_at 1416876_at | 6920 | 0.185 | -0.0516 | No | ||
| 35 | LIMA1 | 1422499_at 1425654_a_at 1435726_at 1435727_s_at 1450629_at | 7040 | 0.180 | -0.0552 | No | ||
| 36 | MAPT | 1417885_at 1424718_at 1424719_a_at 1445634_at 1455028_at | 7076 | 0.179 | -0.0549 | No | ||
| 37 | NRAP | 1421253_at | 7557 | 0.157 | -0.0752 | No | ||
| 38 | ANLN | 1433543_at 1439648_at | 7583 | 0.155 | -0.0747 | No | ||
| 39 | ABI2 | 1433985_at 1436984_at 1444783_at 1458053_at 1460153_at | 7752 | 0.149 | -0.0808 | No | ||
| 40 | ARPC1A | 1416079_a_at | 8119 | 0.136 | -0.0962 | No | ||
| 41 | BIRC5 | 1424278_a_at | 8131 | 0.135 | -0.0952 | No | ||
| 42 | MYO6 | 1421120_at 1433942_at 1435559_at | 8662 | 0.116 | -0.1183 | No | ||
| 43 | CALD1 | 1424768_at 1424769_s_at 1424770_at 1433146_at 1433147_at 1458057_at | 8907 | 0.108 | -0.1284 | No | ||
| 44 | GC | 1426547_at | 8911 | 0.107 | -0.1273 | No | ||
| 45 | APC | 1420956_at 1420957_at 1435543_at 1450056_at | 9040 | 0.103 | -0.1321 | No | ||
| 46 | SHROOM3 | 1422629_s_at 1432377_x_at 1442952_at 1445401_at 1451854_a_at 1453714_a_at 1454211_a_at | 9266 | 0.095 | -0.1414 | No | ||
| 47 | SORBS3 | 1419329_at | 9530 | 0.086 | -0.1526 | No | ||
| 48 | SORBS2 | 1437197_at 1441624_at 1446744_at | 9709 | 0.081 | -0.1599 | No | ||
| 49 | TMOD2 | 1430153_at 1431326_a_at 1435060_at 1437167_at 1439223_at 1451301_at | 9981 | 0.071 | -0.1716 | No | ||
| 50 | NCALD | 1417568_at 1417569_at 1439730_at 1444466_at 1458037_at | 10034 | 0.069 | -0.1732 | No | ||
| 51 | PKD2 | 1417753_at 1441870_s_at | 10236 | 0.063 | -0.1818 | No | ||
| 52 | PDLIM5 | 1429783_at 1438186_at 1442710_at 1446339_at | 10674 | 0.048 | -0.2013 | No | ||
| 53 | CGN | 1430329_at 1435155_at | 10719 | 0.046 | -0.2028 | No | ||
| 54 | DYNC1I1 | 1416361_a_at | 11012 | 0.036 | -0.2158 | No | ||
| 55 | MAP1S | 1454732_at | 11090 | 0.034 | -0.2190 | No | ||
| 56 | RPH3AL | 1431695_at | 11193 | 0.031 | -0.2234 | No | ||
| 57 | IPP | 1421771_a_at | 11226 | 0.029 | -0.2245 | No | ||
| 58 | ACTN2 | 1448327_at 1456968_at | 11955 | 0.005 | -0.2579 | No | ||
| 59 | TNNC1 | 1418370_at | 12393 | -0.011 | -0.2778 | No | ||
| 60 | CCR5 | 1422259_a_at 1422260_x_at 1424727_at | 12496 | -0.015 | -0.2823 | No | ||
| 61 | STMN1 | 1415849_s_at | 12666 | -0.021 | -0.2898 | No | ||
| 62 | TTN | 1427445_a_at 1427446_s_at 1431928_at 1444083_at 1444638_at 1446450_at | 12752 | -0.024 | -0.2935 | No | ||
| 63 | RAB11FIP5 | 1427405_s_at 1434314_s_at | 12789 | -0.026 | -0.2949 | No | ||
| 64 | MYBPC3 | 1418551_at | 12796 | -0.026 | -0.2949 | No | ||
| 65 | MAPK8IP2 | 1418785_at 1418786_at 1435045_s_at 1449225_a_at 1455194_at | 13022 | -0.035 | -0.3048 | No | ||
| 66 | PACSIN3 | 1416995_at 1431534_at 1437834_s_at 1456191_x_at | 13478 | -0.052 | -0.3251 | No | ||
| 67 | DST | 1421117_at 1421276_a_at 1423626_at 1442395_at 1446943_at 1450119_at 1458075_at 1459098_at 1459356_at | 13514 | -0.053 | -0.3262 | No | ||
| 68 | ESPN | 1423005_a_at 1458260_at | 13753 | -0.062 | -0.3364 | No | ||
| 69 | COTL1 | 1416001_a_at 1416002_x_at 1425801_x_at 1436236_x_at 1436838_x_at 1437811_x_at | 13968 | -0.071 | -0.3455 | No | ||
| 70 | ANK1 | 1419421_at 1425677_a_at 1443291_at 1450627_at 1452512_a_at | 14053 | -0.075 | -0.3485 | No | ||
| 71 | RHAG | 1419014_at | 14060 | -0.075 | -0.3480 | No | ||
| 72 | TMOD4 | 1449969_at | 14106 | -0.078 | -0.3492 | No | ||
| 73 | KIF1B | 1423994_at 1423995_at 1425270_at 1451200_at 1451642_at 1451762_a_at 1455182_at 1458426_at | 14744 | -0.106 | -0.3773 | No | ||
| 74 | NEXN | 1435649_at 1446084_at | 14773 | -0.107 | -0.3775 | No | ||
| 75 | BAIAP2 | 1425656_a_at 1435128_at 1451027_at 1451028_at | 14898 | -0.112 | -0.3820 | No | ||
| 76 | ARHGEF2 | 1421042_at 1421043_s_at 1427646_a_at | 15138 | -0.123 | -0.3916 | No | ||
| 77 | DCX | 1418139_at 1418140_at 1418141_at 1448974_at | 15267 | -0.130 | -0.3961 | No | ||
| 78 | CYFIP1 | 1416329_at 1445327_at 1457134_at 1457615_at | 15708 | -0.154 | -0.4147 | No | ||
| 79 | CNN2 | 1450981_at | 15715 | -0.154 | -0.4133 | No | ||
| 80 | SORBS1 | 1417358_s_at 1425826_a_at 1428471_at 1436737_a_at 1440311_at 1442401_at 1443983_at 1455967_at | 15723 | -0.154 | -0.4120 | No | ||
| 81 | SYNPO | 1427045_at 1434089_at | 15940 | -0.166 | -0.4201 | No | ||
| 82 | RHCG | 1417362_at | 15957 | -0.167 | -0.4191 | No | ||
| 83 | ATG4C | 1434014_at 1441440_at | 16401 | -0.196 | -0.4373 | No | ||
| 84 | VCL | 1416156_at 1416157_at 1445256_at | 16767 | -0.221 | -0.4517 | No | ||
| 85 | CROCC | 1427338_at | 16942 | -0.235 | -0.4572 | No | ||
| 86 | SPTBN4 | 1425116_a_at | 16951 | -0.236 | -0.4550 | No | ||
| 87 | TUBGCP3 | 1426707_at | 16961 | -0.236 | -0.4529 | No | ||
| 88 | ARL8A | 1424640_at | 16992 | -0.239 | -0.4518 | No | ||
| 89 | ARPC4 | 1423588_at 1423589_at | 17010 | -0.240 | -0.4500 | No | ||
| 90 | MEFV | 1460283_at | 17030 | -0.242 | -0.4483 | No | ||
| 91 | PXN | 1424027_at 1426085_a_at 1456135_s_at | 17166 | -0.253 | -0.4518 | No | ||
| 92 | DBNL | 1460334_at | 17553 | -0.289 | -0.4664 | No | ||
| 93 | ARFGEF1 | 1415711_at 1437104_at 1444175_at 1456917_at | 17633 | -0.297 | -0.4669 | No | ||
| 94 | MAPK8IP1 | 1425679_a_at 1440619_at | 17822 | -0.318 | -0.4721 | No | ||
| 95 | SSH2 | 1456153_at | 17833 | -0.319 | -0.4692 | No | ||
| 96 | GABARAPL2 | 1423187_at 1441376_at 1444956_at 1459477_at | 17957 | -0.335 | -0.4712 | No | ||
| 97 | MYOZ1 | 1448636_at 1460202_at | 18559 | -0.423 | -0.4943 | No | ||
| 98 | MARCKS | 1415971_at 1415972_at 1430311_at 1437034_x_at 1456028_x_at | 18669 | -0.443 | -0.4946 | No | ||
| 99 | SCIN | 1450276_a_at | 19091 | -0.525 | -0.5083 | No | ||
| 100 | ADD2 | 1421975_a_at 1435287_at 1451914_a_at | 19301 | -0.569 | -0.5118 | Yes | ||
| 101 | CXCR4 | 1448710_at | 19390 | -0.587 | -0.5096 | Yes | ||
| 102 | RABGAP1 | 1424188_at 1441217_at 1441800_at 1443535_at 1457370_at 1460486_at | 19455 | -0.601 | -0.5062 | Yes | ||
| 103 | GABARAP | 1416937_at | 19548 | -0.625 | -0.5037 | Yes | ||
| 104 | ACTA1 | 1427735_a_at | 19700 | -0.666 | -0.5035 | Yes | ||
| 105 | TUBGCP6 | 1435090_at | 19778 | -0.689 | -0.4997 | Yes | ||
| 106 | MAPRE1 | 1422764_at 1422765_at 1428819_at 1428820_at 1450740_a_at | 19895 | -0.725 | -0.4973 | Yes | ||
| 107 | JUP | 1426873_s_at | 19903 | -0.729 | -0.4899 | Yes | ||
| 108 | FLNA | 1426677_at | 19992 | -0.758 | -0.4858 | Yes | ||
| 109 | PALLD | 1427228_at 1433768_at 1440635_at 1440693_at | 20057 | -0.778 | -0.4805 | Yes | ||
| 110 | CLASP2 | 1424836_a_at 1427328_a_at 1429976_at 1440802_at 1441726_at 1443320_at 1456911_at | 20084 | -0.792 | -0.4732 | Yes | ||
| 111 | MARK4 | 1439051_a_at 1456618_at | 20120 | -0.803 | -0.4663 | Yes | ||
| 112 | MYH10 | 1441057_at 1452740_at | 20198 | -0.835 | -0.4609 | Yes | ||
| 113 | TMOD3 | 1423088_at 1423089_at 1438556_a_at 1439626_at 1455708_at 1456913_at | 20361 | -0.908 | -0.4587 | Yes | ||
| 114 | IQGAP2 | 1433885_at 1459894_at | 20368 | -0.912 | -0.4493 | Yes | ||
| 115 | PRNP | 1416130_at 1446507_at 1448233_at | 20440 | -0.945 | -0.4424 | Yes | ||
| 116 | SMTN | 1452469_a_at | 20491 | -0.971 | -0.4344 | Yes | ||
| 117 | DMD | 1417307_at 1430320_at 1446156_at 1448665_at 1457022_at | 20709 | -1.130 | -0.4323 | Yes | ||
| 118 | CDK5RAP2 | 1443976_at 1447103_at 1452618_at | 20728 | -1.144 | -0.4210 | Yes | ||
| 119 | WASF2 | 1438683_at | 20744 | -1.153 | -0.4094 | Yes | ||
| 120 | MYLIP | 1424988_at | 20830 | -1.211 | -0.4004 | Yes | ||
| 121 | KPTN | 1423438_at 1440025_at | 20900 | -1.264 | -0.3901 | Yes | ||
| 122 | TLN1 | 1436042_at 1448402_at 1457782_at | 20979 | -1.328 | -0.3795 | Yes | ||
| 123 | CDC42EP3 | 1422642_at 1450700_at | 21230 | -1.604 | -0.3739 | Yes | ||
| 124 | POLB | 1425371_at 1434229_a_at | 21264 | -1.646 | -0.3578 | Yes | ||
| 125 | ACTN4 | 1423449_a_at 1444258_at | 21337 | -1.743 | -0.3426 | Yes | ||
| 126 | CLASP1 | 1427353_at 1452265_at 1459233_at 1460102_at | 21370 | -1.793 | -0.3250 | Yes | ||
| 127 | MTSS1 | 1424826_s_at 1434036_at 1440847_at 1443729_at 1446284_at 1451496_at | 21390 | -1.847 | -0.3062 | Yes | ||
| 128 | CENPJ | 1423140_at 1423141_at 1437643_at 1450872_s_at | 21410 | -1.876 | -0.2870 | Yes | ||
| 129 | MYO9B | 1418031_at 1438533_at 1459110_at | 21545 | -2.156 | -0.2702 | Yes | ||
| 130 | ATG4D | 1433918_at 1439130_at | 21552 | -2.170 | -0.2474 | Yes | ||
| 131 | KIF5B | 1418427_at 1418428_at 1418429_at 1418430_at 1418431_at | 21625 | -2.334 | -0.2258 | Yes | ||
| 132 | SYNE1 | 1421545_a_at 1455225_at 1455493_at | 21635 | -2.359 | -0.2011 | Yes | ||
| 133 | LSP1 | 1417756_a_at | 21755 | -2.829 | -0.1764 | Yes | ||
| 134 | RAE1 | 1429528_at 1444256_at 1459822_at | 21824 | -3.273 | -0.1447 | Yes | ||
| 135 | UNC84B | 1433832_at 1439238_at | 21879 | -3.868 | -0.1060 | Yes | ||
| 136 | GABARAPL1 | 1416418_at 1416419_s_at 1442572_at | 21881 | -3.884 | -0.0646 | Yes | ||
| 137 | RACGAP1 | 1421546_a_at 1441889_x_at 1451358_a_at | 21926 | -6.293 | 0.0004 | Yes |