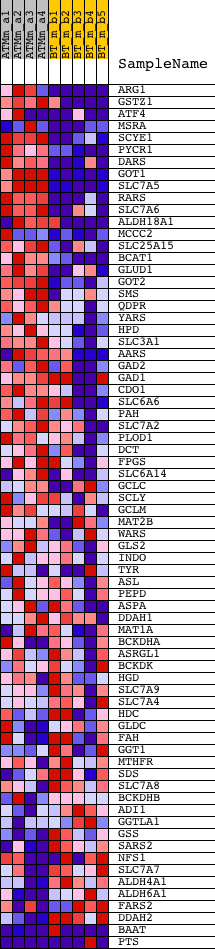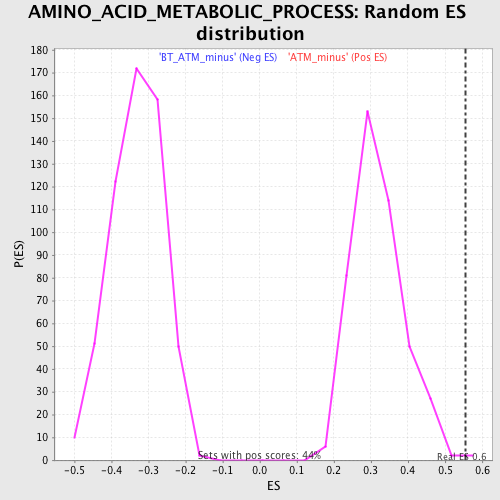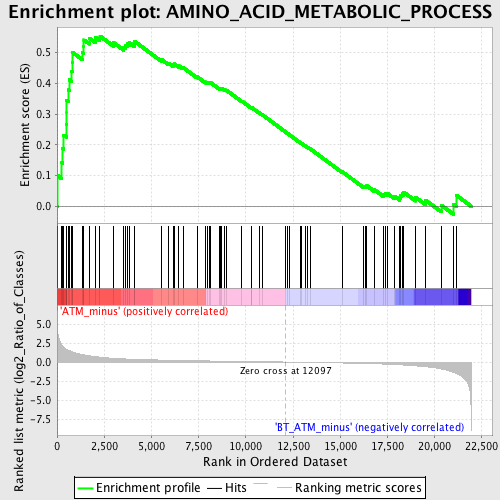
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | AMINO_ACID_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.55393636 |
| Normalized Enrichment Score (NES) | 1.7379873 |
| Nominal p-value | 0.004597701 |
| FDR q-value | 0.2729788 |
| FWER p-Value | 0.813 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARG1 | 1419549_at | 26 | 4.232 | 0.1000 | Yes | ||
| 2 | GSTZ1 | 1427552_a_at 1458134_at | 251 | 2.185 | 0.1420 | Yes | ||
| 3 | ATF4 | 1438992_x_at 1439258_at 1448135_at | 301 | 2.015 | 0.1880 | Yes | ||
| 4 | MSRA | 1442385_at 1448856_a_at 1458923_at | 342 | 1.919 | 0.2321 | Yes | ||
| 5 | SCYE1 | 1416486_at | 490 | 1.686 | 0.2657 | Yes | ||
| 6 | PYCR1 | 1424556_at | 497 | 1.677 | 0.3055 | Yes | ||
| 7 | DARS | 1423800_at | 499 | 1.674 | 0.3455 | Yes | ||
| 8 | GOT1 | 1450970_at | 586 | 1.560 | 0.3789 | Yes | ||
| 9 | SLC7A5 | 1418326_at | 640 | 1.495 | 0.4122 | Yes | ||
| 10 | RARS | 1416312_at | 763 | 1.376 | 0.4395 | Yes | ||
| 11 | SLC7A6 | 1433467_at 1460541_at | 805 | 1.339 | 0.4697 | Yes | ||
| 12 | ALDH18A1 | 1415836_at 1435855_x_at 1437325_x_at 1437333_x_at 1437620_x_at | 812 | 1.330 | 0.5012 | Yes | ||
| 13 | MCCC2 | 1428021_at 1432472_a_at 1454840_at | 1327 | 0.972 | 0.5009 | Yes | ||
| 14 | SLC25A15 | 1420966_at 1420967_at | 1416 | 0.933 | 0.5192 | Yes | ||
| 15 | BCAT1 | 1430111_a_at 1450871_a_at | 1421 | 0.930 | 0.5413 | Yes | ||
| 16 | GLUD1 | 1416209_at 1448253_at | 1710 | 0.807 | 0.5474 | Yes | ||
| 17 | GOT2 | 1417715_a_at 1417716_at | 2043 | 0.701 | 0.5490 | Yes | ||
| 18 | SMS | 1428699_at 1434190_at | 2270 | 0.637 | 0.5539 | Yes | ||
| 19 | QDPR | 1423664_at 1437993_x_at | 2986 | 0.487 | 0.5329 | No | ||
| 20 | YARS | 1460638_at | 3532 | 0.412 | 0.5178 | No | ||
| 21 | HPD | 1424618_at | 3634 | 0.400 | 0.5227 | No | ||
| 22 | SLC3A1 | 1448741_at | 3710 | 0.391 | 0.5287 | No | ||
| 23 | AARS | 1423685_at 1451083_s_at | 3834 | 0.379 | 0.5321 | No | ||
| 24 | GAD2 | 1421978_at | 4105 | 0.356 | 0.5283 | No | ||
| 25 | GAD1 | 1416561_at 1416562_at | 4113 | 0.356 | 0.5365 | No | ||
| 26 | CDO1 | 1448842_at | 5531 | 0.257 | 0.4778 | No | ||
| 27 | SLC6A6 | 1420148_at 1421346_a_at 1437149_at 1449751_at | 5918 | 0.234 | 0.4657 | No | ||
| 28 | PAH | 1454638_a_at | 6143 | 0.222 | 0.4608 | No | ||
| 29 | SLC7A2 | 1422648_at 1426008_a_at 1436555_at 1440506_at 1450703_at | 6198 | 0.219 | 0.4636 | No | ||
| 30 | PLOD1 | 1416289_at 1445893_at | 6453 | 0.207 | 0.4569 | No | ||
| 31 | DCT | 1418028_at | 6666 | 0.197 | 0.4519 | No | ||
| 32 | FPGS | 1460673_at | 7417 | 0.163 | 0.4215 | No | ||
| 33 | SLC6A14 | 1420503_at 1420504_at | 7860 | 0.146 | 0.4048 | No | ||
| 34 | GCLC | 1424296_at | 7979 | 0.141 | 0.4027 | No | ||
| 35 | SCLY | 1417671_at 1441192_at | 8069 | 0.137 | 0.4019 | No | ||
| 36 | GCLM | 1418627_at | 8114 | 0.136 | 0.4032 | No | ||
| 37 | MAT2B | 1448196_at | 8600 | 0.118 | 0.3838 | No | ||
| 38 | WARS | 1415694_at 1425106_a_at 1434813_x_at 1437832_x_at | 8670 | 0.116 | 0.3835 | No | ||
| 39 | GLS2 | 1435245_at | 8728 | 0.114 | 0.3836 | No | ||
| 40 | INDO | 1420437_at | 8887 | 0.108 | 0.3789 | No | ||
| 41 | TYR | 1417717_a_at 1448821_at 1456095_at | 8967 | 0.106 | 0.3779 | No | ||
| 42 | ASL | 1448350_at | 9785 | 0.078 | 0.3424 | No | ||
| 43 | PEPD | 1416712_at | 10281 | 0.061 | 0.3212 | No | ||
| 44 | ASPA | 1418472_at | 10297 | 0.061 | 0.3219 | No | ||
| 45 | DDAH1 | 1429298_at 1429299_at 1434519_at 1438879_at 1454995_at 1455400_at | 10709 | 0.046 | 0.3042 | No | ||
| 46 | MAT1A | 1423147_at | 10858 | 0.042 | 0.2985 | No | ||
| 47 | BCKDHA | 1416647_at | 12105 | -0.000 | 0.2415 | No | ||
| 48 | ASRGL1 | 1424395_at 1424396_a_at 1460360_at | 12180 | -0.003 | 0.2382 | No | ||
| 49 | BCKDK | 1443813_x_at 1460644_at | 12298 | -0.007 | 0.2330 | No | ||
| 50 | HGD | 1452986_at | 12887 | -0.029 | 0.2068 | No | ||
| 51 | SLC7A9 | 1448783_at | 12952 | -0.032 | 0.2047 | No | ||
| 52 | SLC7A4 | 1426068_at 1426069_s_at 1436776_x_at | 13173 | -0.040 | 0.1956 | No | ||
| 53 | HDC | 1451796_s_at 1454713_s_at | 13284 | -0.044 | 0.1916 | No | ||
| 54 | GLDC | 1416049_at 1459959_at | 13399 | -0.049 | 0.1875 | No | ||
| 55 | FAH | 1417220_at | 15089 | -0.120 | 0.1131 | No | ||
| 56 | GGT1 | 1448485_at | 16244 | -0.185 | 0.0648 | No | ||
| 57 | MTHFR | 1422132_at 1434087_at 1450498_at | 16341 | -0.191 | 0.0650 | No | ||
| 58 | SDS | 1424744_at | 16402 | -0.196 | 0.0669 | No | ||
| 59 | SLC7A8 | 1417929_at | 16800 | -0.224 | 0.0541 | No | ||
| 60 | BCKDHB | 1427153_at | 17282 | -0.263 | 0.0384 | No | ||
| 61 | ADI1 | 1449076_x_at | 17370 | -0.271 | 0.0409 | No | ||
| 62 | GGTLA1 | 1418216_at 1439420_x_at 1455747_at | 17471 | -0.280 | 0.0430 | No | ||
| 63 | GSS | 1441931_x_at 1448273_at | 17883 | -0.325 | 0.0320 | No | ||
| 64 | SARS2 | 1448522_at | 18152 | -0.358 | 0.0283 | No | ||
| 65 | NFS1 | 1416373_at 1431431_a_at 1442017_at | 18194 | -0.365 | 0.0351 | No | ||
| 66 | SLC7A7 | 1417392_a_at 1447181_s_at | 18266 | -0.376 | 0.0409 | No | ||
| 67 | ALDH4A1 | 1452375_at | 18361 | -0.391 | 0.0459 | No | ||
| 68 | ALDH6A1 | 1448104_at | 18987 | -0.507 | 0.0295 | No | ||
| 69 | FARS2 | 1431354_a_at 1439406_x_at | 19507 | -0.612 | 0.0204 | No | ||
| 70 | DDAH2 | 1416457_at | 20354 | -0.905 | 0.0033 | No | ||
| 71 | BAAT | 1419001_at 1419002_s_at | 21007 | -1.365 | 0.0062 | No | ||
| 72 | PTS | 1450660_at 1458402_at | 21142 | -1.514 | 0.0362 | No |