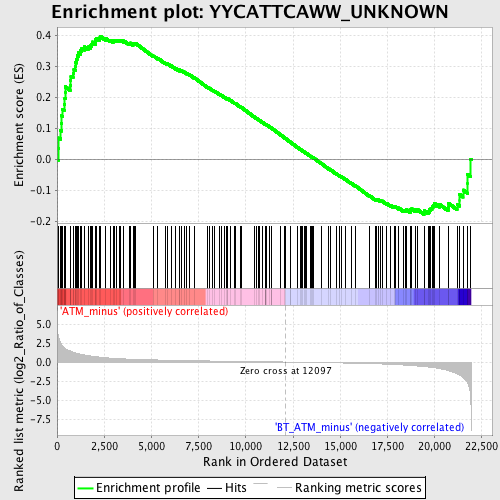
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | YYCATTCAWW_UNKNOWN |
| Enrichment Score (ES) | 0.3981132 |
| Normalized Enrichment Score (NES) | 1.4011656 |
| Nominal p-value | 0.011574074 |
| FDR q-value | 0.4360029 |
| FWER p-Value | 0.985 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TOMM40 | 1426118_a_at 1459680_at | 54 | 3.547 | 0.0370 | Yes | ||
| 2 | BMP7 | 1418910_at 1432410_a_at | 93 | 3.103 | 0.0698 | Yes | ||
| 3 | XPO1 | 1418442_at 1418443_at 1448070_at | 176 | 2.542 | 0.0944 | Yes | ||
| 4 | NR4A3 | 1421079_at 1421080_at | 228 | 2.289 | 0.1175 | Yes | ||
| 5 | CYR61 | 1416039_x_at 1417848_at 1417849_at 1438133_a_at 1442340_x_at 1457823_at 1459462_at | 248 | 2.191 | 0.1411 | Yes | ||
| 6 | RGS3 | 1425701_a_at 1427648_at 1449516_a_at 1454026_a_at | 297 | 2.018 | 0.1613 | Yes | ||
| 7 | NRK | 1436399_s_at 1450078_at 1450079_at | 373 | 1.858 | 0.1786 | Yes | ||
| 8 | PAPPA | 1427633_a_at 1432591_at 1432592_at 1459375_at | 412 | 1.793 | 0.1968 | Yes | ||
| 9 | TIAL1 | 1421148_a_at 1426352_s_at 1446713_at 1452821_at 1455675_a_at 1455676_x_at | 430 | 1.763 | 0.2157 | Yes | ||
| 10 | EEF1G | 1417364_at 1458955_at | 438 | 1.751 | 0.2349 | Yes | ||
| 11 | PGM3 | 1428228_at | 689 | 1.451 | 0.2395 | Yes | ||
| 12 | ATP5G1 | 1416020_a_at 1444874_at | 708 | 1.428 | 0.2546 | Yes | ||
| 13 | ERBB4 | 1427783_at 1446211_at 1447043_at | 733 | 1.404 | 0.2692 | Yes | ||
| 14 | EBAG9 | 1422669_at | 846 | 1.299 | 0.2785 | Yes | ||
| 15 | CLTC | 1419407_at 1439094_at 1440457_at 1454626_at | 891 | 1.258 | 0.2905 | Yes | ||
| 16 | AIM1L | 1437813_at | 960 | 1.207 | 0.3008 | Yes | ||
| 17 | ETV5 | 1420998_at 1428142_at 1450082_s_at | 973 | 1.197 | 0.3136 | Yes | ||
| 18 | TSC1 | 1422043_at 1439989_at 1455252_at | 1023 | 1.168 | 0.3244 | Yes | ||
| 19 | P4HA1 | 1426519_at 1452094_at | 1060 | 1.142 | 0.3354 | Yes | ||
| 20 | ID1 | 1425895_a_at | 1130 | 1.099 | 0.3445 | Yes | ||
| 21 | PPM1A | 1415678_at 1417221_at 1425537_at 1429500_at 1429501_s_at 1443903_at 1451943_a_at 1453171_s_at | 1247 | 1.028 | 0.3506 | Yes | ||
| 22 | KLF12 | 1439846_at 1439847_s_at 1441040_at 1455521_at | 1309 | 0.984 | 0.3588 | Yes | ||
| 23 | VDAC2 | 1415990_at | 1433 | 0.926 | 0.3635 | Yes | ||
| 24 | COX8A | 1416112_at 1436409_at 1448222_x_at | 1642 | 0.842 | 0.3633 | Yes | ||
| 25 | RANBP1 | 1422547_at | 1763 | 0.792 | 0.3666 | Yes | ||
| 26 | CDC40 | 1442667_at 1445348_at 1453342_at 1457366_at | 1823 | 0.769 | 0.3725 | Yes | ||
| 27 | LRP8 | 1421459_a_at 1440882_at 1442347_at | 1860 | 0.756 | 0.3792 | Yes | ||
| 28 | E2F3 | 1427462_at 1434564_at | 2034 | 0.705 | 0.3791 | Yes | ||
| 29 | DLX1 | 1449470_at | 2041 | 0.702 | 0.3867 | Yes | ||
| 30 | UBE2N | 1422559_at 1435384_at | 2100 | 0.683 | 0.3916 | Yes | ||
| 31 | ETV4 | 1423232_at 1443381_at | 2245 | 0.642 | 0.3922 | Yes | ||
| 32 | EPHB2 | 1425015_at 1425016_at 1442471_at 1445417_at 1454022_at | 2271 | 0.636 | 0.3981 | Yes | ||
| 33 | CCND1 | 1417419_at 1417420_at 1448698_at | 2582 | 0.561 | 0.3901 | No | ||
| 34 | FOSB | 1422134_at | 2811 | 0.516 | 0.3854 | No | ||
| 35 | DCAMKL1 | 1423125_at 1424270_at 1424271_at 1436659_at 1446190_at 1450863_a_at 1451289_at 1451917_a_at | 2966 | 0.490 | 0.3838 | No | ||
| 36 | ATF3 | 1449363_at | 3055 | 0.476 | 0.3851 | No | ||
| 37 | DUSP4 | 1428834_at | 3159 | 0.462 | 0.3855 | No | ||
| 38 | TPM3 | 1427260_a_at 1427567_a_at 1436958_x_at 1449996_a_at 1449997_at | 3296 | 0.442 | 0.3842 | No | ||
| 39 | LGALS3 | 1426808_at 1445626_at | 3374 | 0.432 | 0.3854 | No | ||
| 40 | LOXL4 | 1421153_at 1450134_at | 3534 | 0.412 | 0.3827 | No | ||
| 41 | TRIB2 | 1426640_s_at 1426641_at 1459852_x_at | 3808 | 0.381 | 0.3744 | No | ||
| 42 | SCG2 | 1450708_at | 3866 | 0.375 | 0.3760 | No | ||
| 43 | TNFSF11 | 1419083_at 1451944_a_at | 4020 | 0.364 | 0.3730 | No | ||
| 44 | MXI1 | 1425732_a_at 1450376_at | 4082 | 0.359 | 0.3742 | No | ||
| 45 | ZDHHC21 | 1429488_at | 4149 | 0.351 | 0.3751 | No | ||
| 46 | RWDD2 | 1428963_at | 5112 | 0.285 | 0.3341 | No | ||
| 47 | MYO1C | 1419648_at 1419649_s_at 1449550_at 1449551_at | 5331 | 0.270 | 0.3271 | No | ||
| 48 | PIP5K2B | 1427438_at 1435068_at 1441164_at 1444128_at | 5740 | 0.244 | 0.3111 | No | ||
| 49 | GPM6A | 1426442_at 1456741_s_at 1459303_at | 5843 | 0.238 | 0.3091 | No | ||
| 50 | RAB30 | 1426452_a_at | 6043 | 0.227 | 0.3025 | No | ||
| 51 | CSPG2 | 1421694_a_at 1427256_at 1427257_at 1433043_at 1433044_at 1447586_at 1447887_x_at 1459767_x_at | 6293 | 0.215 | 0.2934 | No | ||
| 52 | EFEMP2 | 1417018_at 1447668_x_at | 6470 | 0.206 | 0.2877 | No | ||
| 53 | NEDD4 | 1421955_a_at 1444044_at 1450431_a_at 1451109_a_at | 6483 | 0.206 | 0.2894 | No | ||
| 54 | AGTR2 | 1415832_at | 6574 | 0.201 | 0.2875 | No | ||
| 55 | GGN | 1445278_at | 6730 | 0.193 | 0.2825 | No | ||
| 56 | GAP43 | 1423537_at 1445567_at | 6876 | 0.187 | 0.2780 | No | ||
| 57 | DNM1L | 1428008_at 1428086_at 1428087_at 1452638_s_at | 7010 | 0.182 | 0.2739 | No | ||
| 58 | NF2 | 1421820_a_at 1427708_a_at 1450382_at 1451829_a_at | 7252 | 0.170 | 0.2647 | No | ||
| 59 | GCLC | 1424296_at | 7979 | 0.141 | 0.2330 | No | ||
| 60 | RHEBL1 | 1447818_x_at 1451516_at | 8058 | 0.138 | 0.2309 | No | ||
| 61 | RAPH1 | 1434302_at 1434303_at 1444586_at 1456918_at | 8242 | 0.132 | 0.2240 | No | ||
| 62 | KIAA1128 | 1431972_a_at 1452223_s_at 1453240_a_at | 8339 | 0.128 | 0.2210 | No | ||
| 63 | TCF15 | 1449592_at | 8597 | 0.119 | 0.2105 | No | ||
| 64 | KCNQ5 | 1427426_at 1441071_at | 8715 | 0.115 | 0.2064 | No | ||
| 65 | FOXD3 | 1422210_at | 8848 | 0.110 | 0.2016 | No | ||
| 66 | LMNB1 | 1423520_at 1423521_at | 8986 | 0.105 | 0.1965 | No | ||
| 67 | DIO2 | 1418937_at 1418938_at 1426081_a_at | 9015 | 0.104 | 0.1963 | No | ||
| 68 | AAK1 | 1420026_at 1435038_s_at 1441782_at | 9045 | 0.103 | 0.1962 | No | ||
| 69 | RBPSUH | 1418114_at 1438848_at 1446436_at 1448957_at 1454896_at | 9180 | 0.098 | 0.1911 | No | ||
| 70 | FGFR3 | 1421841_at 1425796_a_at | 9407 | 0.090 | 0.1817 | No | ||
| 71 | HTF9C | 1434244_x_at 1441920_x_at 1448114_a_at 1448115_at 1453995_a_at 1457620_at | 9426 | 0.090 | 0.1819 | No | ||
| 72 | DDR2 | 1422738_at 1455688_at | 9727 | 0.080 | 0.1690 | No | ||
| 73 | DNAJB5 | 1421961_a_at 1421962_at 1450436_s_at | 9783 | 0.078 | 0.1674 | No | ||
| 74 | GPR3 | 1460275_at | 10445 | 0.056 | 0.1377 | No | ||
| 75 | LIF | 1421206_at 1421207_at 1450160_at | 10555 | 0.052 | 0.1332 | No | ||
| 76 | PTPN11 | 1421196_at 1427699_a_at 1451225_at | 10663 | 0.048 | 0.1289 | No | ||
| 77 | FREQ | 1434887_at 1450146_at 1460293_at | 10710 | 0.046 | 0.1273 | No | ||
| 78 | SCN3A | 1421705_at 1439204_at | 10895 | 0.040 | 0.1193 | No | ||
| 79 | CPS1 | 1455540_at | 11011 | 0.036 | 0.1144 | No | ||
| 80 | NUP98 | 1434220_at 1439154_at 1446397_at 1446988_at | 11029 | 0.036 | 0.1140 | No | ||
| 81 | IGF1R | 1426565_at 1437219_at 1443566_at 1445600_at 1445664_at 1446303_at | 11032 | 0.036 | 0.1143 | No | ||
| 82 | USP7 | 1419920_s_at 1419921_s_at 1434371_x_at 1437118_at 1438249_at 1454948_at 1454949_at | 11040 | 0.036 | 0.1144 | No | ||
| 83 | NEUROD6 | 1418047_at | 11064 | 0.035 | 0.1137 | No | ||
| 84 | FILIP1 | 1436650_at 1444075_at | 11264 | 0.028 | 0.1049 | No | ||
| 85 | PLA2G3 | 1441247_at 1456231_at | 11328 | 0.026 | 0.1023 | No | ||
| 86 | ANGPT2 | 1422415_at 1446903_at 1448831_at | 11351 | 0.026 | 0.1016 | No | ||
| 87 | VAX1 | 1421774_at | 11354 | 0.025 | 0.1018 | No | ||
| 88 | INHBA | 1422053_at 1458291_at | 11826 | 0.009 | 0.0803 | No | ||
| 89 | BRMS1 | 1417897_at 1426129_at | 12063 | 0.001 | 0.0694 | No | ||
| 90 | EIF4E | 1450909_at 1457489_at 1459371_at | 12114 | -0.001 | 0.0672 | No | ||
| 91 | ANK3 | 1425202_a_at 1439220_at 1447259_at 1451628_a_at 1457288_at | 12365 | -0.010 | 0.0558 | No | ||
| 92 | CDC7 | 1426002_a_at 1426021_a_at | 12379 | -0.011 | 0.0553 | No | ||
| 93 | SCG3 | 1448628_at | 12715 | -0.023 | 0.0402 | No | ||
| 94 | SALL3 | 1441186_at 1452386_at | 12884 | -0.029 | 0.0328 | No | ||
| 95 | MCC | 1438081_at 1442741_at 1442892_at 1459524_at 1459752_at | 12963 | -0.033 | 0.0296 | No | ||
| 96 | PPAP2B | 1429514_at 1446850_at 1448908_at | 12982 | -0.033 | 0.0291 | No | ||
| 97 | FOSL1 | 1417487_at 1417488_at | 13122 | -0.038 | 0.0232 | No | ||
| 98 | MYH2 | 1425153_at | 13172 | -0.040 | 0.0214 | No | ||
| 99 | NFIA | 1421162_a_at 1421163_a_at 1427733_a_at 1438236_at 1440660_at 1441547_at 1446742_at 1446990_at 1456087_at 1459431_at | 13204 | -0.041 | 0.0204 | No | ||
| 100 | USP10 | 1448230_at | 13420 | -0.049 | 0.0111 | No | ||
| 101 | LAMA1 | 1418153_at | 13475 | -0.052 | 0.0092 | No | ||
| 102 | ACCN1 | 1417994_a_at | 13520 | -0.054 | 0.0078 | No | ||
| 103 | GABRA1 | 1421280_at 1421281_at 1436889_at 1455766_at | 13587 | -0.056 | 0.0054 | No | ||
| 104 | USP14 | 1416208_at 1437714_x_at 1439201_at 1455829_at | 13975 | -0.072 | -0.0116 | No | ||
| 105 | KATNB1 | 1426665_at | 14392 | -0.089 | -0.0297 | No | ||
| 106 | B3GNT6 | 1456680_at | 14494 | -0.094 | -0.0333 | No | ||
| 107 | SENP8 | 1437307_at 1438102_at 1454084_a_at | 14795 | -0.108 | -0.0459 | No | ||
| 108 | DAAM1 | 1431035_at 1455244_at 1458662_at | 14955 | -0.115 | -0.0519 | No | ||
| 109 | HCRTR1 | 1436295_at | 15059 | -0.119 | -0.0553 | No | ||
| 110 | RECQL | 1418339_at 1425798_a_at 1436371_at 1439047_s_at 1447198_at | 15251 | -0.129 | -0.0626 | No | ||
| 111 | CASK | 1422518_at 1422519_at 1427692_a_at 1445152_at | 15564 | -0.145 | -0.0753 | No | ||
| 112 | CALM1 | 1417365_a_at 1417366_s_at 1433592_at 1454611_a_at 1455571_x_at | 15802 | -0.158 | -0.0844 | No | ||
| 113 | GALE | 1424140_at | 16553 | -0.206 | -0.1166 | No | ||
| 114 | SLC26A7 | 1425841_at | 16866 | -0.229 | -0.1283 | No | ||
| 115 | ADNP | 1423069_at | 16939 | -0.234 | -0.1290 | No | ||
| 116 | WASF1 | 1418545_at 1440438_at | 17003 | -0.240 | -0.1293 | No | ||
| 117 | CDC14A | 1436913_at 1443184_at 1446493_at 1459517_at | 17119 | -0.249 | -0.1318 | No | ||
| 118 | RFX4 | 1432053_at 1436931_at 1443312_at | 17214 | -0.257 | -0.1332 | No | ||
| 119 | TUBA3 | 1438611_at | 17454 | -0.278 | -0.1411 | No | ||
| 120 | ELOVL6 | 1417403_at 1417404_at 1445062_at 1445578_at 1445641_at | 17659 | -0.300 | -0.1471 | No | ||
| 121 | CHD2 | 1444246_at 1445843_at | 17842 | -0.321 | -0.1519 | No | ||
| 122 | RAB35 | 1433922_at | 17900 | -0.327 | -0.1509 | No | ||
| 123 | SP4 | 1421504_at 1437508_at | 18088 | -0.350 | -0.1555 | No | ||
| 124 | AP1B1 | 1416279_at | 18364 | -0.392 | -0.1638 | No | ||
| 125 | PRKAR2B | 1430640_a_at 1438664_at 1456475_s_at | 18440 | -0.403 | -0.1627 | No | ||
| 126 | SPRY4 | 1422020_at 1422021_at 1440867_at 1445669_at | 18486 | -0.412 | -0.1602 | No | ||
| 127 | PLCB2 | 1452481_at | 18712 | -0.449 | -0.1656 | No | ||
| 128 | MAFF | 1418936_at | 18727 | -0.452 | -0.1612 | No | ||
| 129 | BASP1 | 1428572_at | 18789 | -0.465 | -0.1588 | No | ||
| 130 | RAB5C | 1424684_at | 18977 | -0.505 | -0.1617 | No | ||
| 131 | DLEU2 | 1427410_at 1427411_s_at 1456145_at | 19079 | -0.524 | -0.1605 | No | ||
| 132 | MYO9A | 1436307_at 1439361_at 1441567_at 1444541_at 1459991_at | 19433 | -0.597 | -0.1701 | No | ||
| 133 | CHD6 | 1427384_at 1431047_at | 19467 | -0.605 | -0.1649 | No | ||
| 134 | CORO1A | 1416246_a_at 1435288_at 1455269_a_at | 19656 | -0.651 | -0.1663 | No | ||
| 135 | EMID1 | 1442144_at 1449581_at | 19719 | -0.671 | -0.1616 | No | ||
| 136 | FLI1 | 1422024_at 1433512_at 1441584_at | 19797 | -0.694 | -0.1574 | No | ||
| 137 | EP300 | 1434765_at | 19856 | -0.714 | -0.1521 | No | ||
| 138 | SMARCA4 | 1426804_at 1426805_at | 19909 | -0.732 | -0.1464 | No | ||
| 139 | DDX5 | 1419653_a_at 1423645_a_at 1433809_at 1433810_x_at 1438371_x_at 1454793_x_at | 19988 | -0.757 | -0.1415 | No | ||
| 140 | FYN | 1417558_at 1441647_at 1448765_at | 20236 | -0.853 | -0.1434 | No | ||
| 141 | DMD | 1417307_at 1430320_at 1446156_at 1448665_at 1457022_at | 20709 | -1.130 | -0.1524 | No | ||
| 142 | UBE2S | 1416726_s_at 1430962_at | 20738 | -1.150 | -0.1409 | No | ||
| 143 | CKB | 1455106_a_at | 21181 | -1.551 | -0.1439 | No | ||
| 144 | DPF1 | 1420529_at 1420530_at 1442440_at | 21307 | -1.694 | -0.1308 | No | ||
| 145 | TOB1 | 1423176_at 1440844_at | 21309 | -1.696 | -0.1120 | No | ||
| 146 | RUNX1 | 1422864_at 1422865_at 1427650_a_at 1427847_at 1440878_at 1444527_at 1446930_at 1452530_a_at 1452531_at 1452578_at 1459470_at | 21496 | -2.039 | -0.0978 | No | ||
| 147 | PDE4B | 1422473_at 1422474_at 1442700_at 1444731_at 1445449_at 1446577_at 1447237_at 1447718_at | 21732 | -2.726 | -0.0782 | No | ||
| 148 | ATF7IP | 1446323_at 1449192_at 1454973_at 1459096_at | 21738 | -2.750 | -0.0478 | No | ||
| 149 | ETS2 | 1416268_at 1437809_x_at 1447685_x_at | 21910 | -5.096 | 0.0011 | No |

