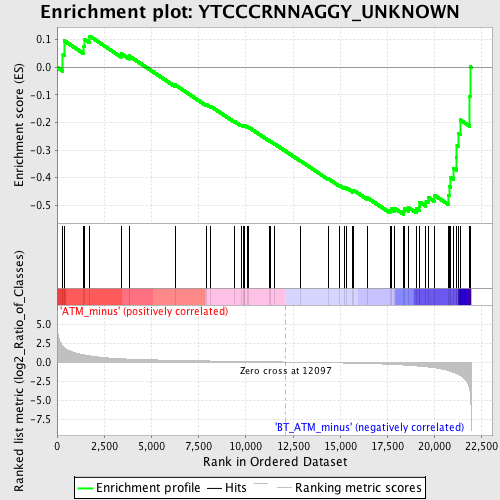
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | YTCCCRNNAGGY_UNKNOWN |
| Enrichment Score (ES) | -0.5333054 |
| Normalized Enrichment Score (NES) | -1.5089339 |
| Nominal p-value | 0.019855596 |
| FDR q-value | 0.14706284 |
| FWER p-Value | 0.893 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SNAPC3 | 1428760_at 1428761_a_at 1453077_a_at 1457731_at | 310 | 1.990 | 0.0446 | No | ||
| 2 | CCT8 | 1415785_a_at 1436973_at | 380 | 1.845 | 0.0958 | No | ||
| 3 | RUNX3 | 1421467_at 1440275_at | 1417 | 0.932 | 0.0760 | No | ||
| 4 | IER3 | 1419647_a_at | 1460 | 0.914 | 0.1010 | No | ||
| 5 | SEPT10 | 1437208_at 1438804_at | 1726 | 0.803 | 0.1126 | No | ||
| 6 | TCEAL1 | 1424634_at | 3385 | 0.431 | 0.0496 | No | ||
| 7 | GSPT1 | 1426736_at 1426737_at 1435000_at 1446550_at 1452168_x_at 1455173_at | 3821 | 0.380 | 0.0409 | No | ||
| 8 | BACH2 | 1421343_at 1437667_a_at 1439468_at 1440304_at 1441657_at 1442605_at 1443263_at 1446929_at 1458730_at 1459136_at | 6262 | 0.216 | -0.0642 | No | ||
| 9 | POLK | 1419501_at 1449483_at | 7889 | 0.145 | -0.1342 | No | ||
| 10 | ADAM11 | 1450248_at | 8123 | 0.136 | -0.1408 | No | ||
| 11 | TGFB3 | 1417455_at | 9412 | 0.090 | -0.1970 | No | ||
| 12 | GSC | 1421412_at | 9749 | 0.079 | -0.2100 | No | ||
| 13 | MITF | 1422025_at 1455214_at | 9875 | 0.075 | -0.2135 | No | ||
| 14 | ZDHHC15 | 1442274_at | 9924 | 0.074 | -0.2136 | No | ||
| 15 | PACRG | 1428751_at | 9936 | 0.073 | -0.2119 | No | ||
| 16 | HOXD8 | 1431099_at 1433638_s_at | 10057 | 0.069 | -0.2154 | No | ||
| 17 | PCDH11X | 1446581_at 1459481_at | 10148 | 0.066 | -0.2175 | No | ||
| 18 | APBA3 | 1416182_at | 11245 | 0.029 | -0.2668 | No | ||
| 19 | OSM | 1438767_at | 11294 | 0.027 | -0.2682 | No | ||
| 20 | GTF3C2 | 1428448_a_at 1428449_at 1440307_at 1452780_at 1452781_a_at | 11525 | 0.020 | -0.2781 | No | ||
| 21 | PPP2R3A | 1455198_a_at | 12886 | -0.029 | -0.3394 | No | ||
| 22 | ZW10 | 1424891_a_at 1431747_at 1445206_at | 14350 | -0.087 | -0.4036 | No | ||
| 23 | MRPL54 | 1448699_at 1456788_at 1457378_at | 14970 | -0.115 | -0.4285 | No | ||
| 24 | IL17RC | 1419671_a_at 1457471_at | 15235 | -0.128 | -0.4368 | No | ||
| 25 | FASTK | 1460635_at | 15331 | -0.132 | -0.4372 | No | ||
| 26 | NTN2L | 1422056_at | 15661 | -0.151 | -0.4478 | No | ||
| 27 | PABPC1 | 1418883_a_at 1419500_at 1453840_at | 15692 | -0.152 | -0.4447 | No | ||
| 28 | HS6ST2 | 1420938_at 1420939_at 1446257_at 1450047_at | 16422 | -0.197 | -0.4722 | No | ||
| 29 | SLC9A1 | 1417397_at | 17632 | -0.297 | -0.5187 | No | ||
| 30 | PTPN23 | 1427128_at | 17706 | -0.305 | -0.5130 | No | ||
| 31 | ORMDL3 | 1419450_at | 17882 | -0.325 | -0.5114 | No | ||
| 32 | CDK9 | 1417269_at 1457240_at | 18362 | -0.391 | -0.5218 | Yes | ||
| 33 | TBX6 | 1449868_at | 18377 | -0.393 | -0.5108 | Yes | ||
| 34 | SNX27 | 1430037_at 1433614_at | 18617 | -0.432 | -0.5090 | Yes | ||
| 35 | SKP2 | 1418969_at 1425072_at 1436000_a_at 1437033_a_at 1449293_a_at 1460247_a_at | 19029 | -0.514 | -0.5126 | Yes | ||
| 36 | HACE1 | 1442163_at 1452278_a_at 1459130_at | 19186 | -0.547 | -0.5036 | Yes | ||
| 37 | RPS6KB2 | 1422268_a_at | 19212 | -0.552 | -0.4884 | Yes | ||
| 38 | PARK2 | 1420755_a_at 1426135_a_at 1449975_a_at | 19535 | -0.623 | -0.4848 | Yes | ||
| 39 | GRWD1 | 1436887_x_at 1452171_at 1455841_s_at | 19651 | -0.650 | -0.4708 | Yes | ||
| 40 | FLOT1 | 1448559_at | 19984 | -0.757 | -0.4637 | Yes | ||
| 41 | IRF2BP1 | 1451252_at | 20712 | -1.132 | -0.4635 | Yes | ||
| 42 | WBP2 | 1448121_at | 20774 | -1.176 | -0.4316 | Yes | ||
| 43 | BCAS3 | 1423528_at 1428454_at 1457817_at 1458557_at | 20844 | -1.220 | -0.3988 | Yes | ||
| 44 | BTBD1 | 1445646_at 1455286_at 1456222_at | 21000 | -1.357 | -0.3658 | Yes | ||
| 45 | COL4A3BP | 1420383_a_at 1420384_at 1449847_a_at 1452867_at | 21166 | -1.529 | -0.3282 | Yes | ||
| 46 | ERO1LB | 1425705_a_at 1434714_at 1449948_at | 21177 | -1.547 | -0.2830 | Yes | ||
| 47 | RDH12 | 1424256_at 1431010_a_at | 21248 | -1.621 | -0.2384 | Yes | ||
| 48 | WHSC1L1 | 1442085_at 1447931_at 1455668_at 1457630_at 1457793_a_at 1457794_at 1459907_a_at | 21356 | -1.776 | -0.1909 | Yes | ||
| 49 | RANBP3 | 1452673_at 1457711_at 1460612_at | 21859 | -3.619 | -0.1070 | Yes | ||
| 50 | ARL6IP2 | 1416793_at 1416794_at 1431197_at 1435594_at | 21871 | -3.742 | 0.0029 | Yes |

