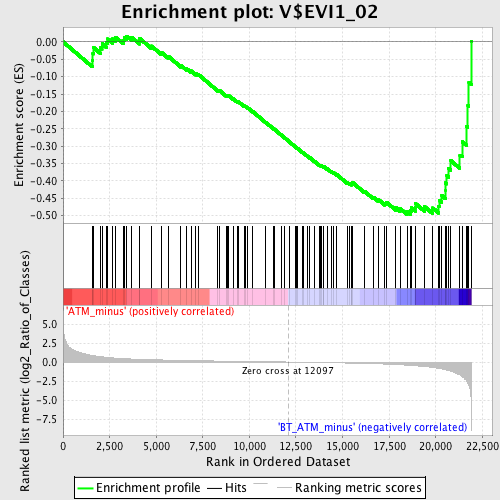
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | V$EVI1_02 |
| Enrichment Score (ES) | -0.49650434 |
| Normalized Enrichment Score (NES) | -1.5723354 |
| Nominal p-value | 0.0063795852 |
| FDR q-value | 0.14130802 |
| FWER p-Value | 0.65 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SDCCAG3 | 1429554_at 1431760_a_at 1453195_at 1456106_x_at | 1570 | 0.864 | -0.0522 | No | ||
| 2 | SYNCRIP | 1422768_at 1422769_at 1426402_at 1428546_at 1450743_s_at | 1599 | 0.855 | -0.0341 | No | ||
| 3 | H3F3A | 1423263_at 1442087_at | 1620 | 0.848 | -0.0157 | No | ||
| 4 | PPARGC1A | 1434099_at 1434100_x_at 1437751_at 1456395_at 1460336_at | 2000 | 0.714 | -0.0168 | No | ||
| 5 | HIPK1 | 1421298_a_at 1424540_at 1457515_at | 2089 | 0.686 | -0.0052 | No | ||
| 6 | SLC7A3 | 1417022_at | 2316 | 0.624 | -0.0014 | No | ||
| 7 | MPDU1 | 1416104_at | 2404 | 0.604 | 0.0083 | No | ||
| 8 | MSX2 | 1438351_at 1449559_at | 2671 | 0.539 | 0.0084 | No | ||
| 9 | SOX11 | 1429051_s_at 1429372_at 1431225_at 1436790_a_at 1453002_at 1453125_at | 2813 | 0.516 | 0.0137 | No | ||
| 10 | CTNNA3 | 1441560_at | 3237 | 0.450 | 0.0046 | No | ||
| 11 | KCNMB2 | 1426322_a_at 1431844_at | 3275 | 0.444 | 0.0130 | No | ||
| 12 | SIX1 | 1427277_at | 3412 | 0.428 | 0.0165 | No | ||
| 13 | PHEX | 1421979_at 1450445_at | 3677 | 0.395 | 0.0134 | No | ||
| 14 | ADPRHL1 | 1455952_at | 4109 | 0.356 | 0.0017 | No | ||
| 15 | GAD1 | 1416561_at 1416562_at | 4113 | 0.356 | 0.0097 | No | ||
| 16 | CAPNS2 | 1429067_at | 4737 | 0.307 | -0.0119 | No | ||
| 17 | HSPB3 | 1449872_at | 5271 | 0.274 | -0.0301 | No | ||
| 18 | NHLH2 | 1436888_at | 5641 | 0.249 | -0.0413 | No | ||
| 19 | HTR3B | 1421652_at | 6329 | 0.213 | -0.0679 | No | ||
| 20 | ADAMTS3 | 1441693_at 1458143_at | 6640 | 0.198 | -0.0776 | No | ||
| 21 | NCAM2 | 1421592_at 1421593_at 1425301_at 1459116_at | 6867 | 0.187 | -0.0837 | No | ||
| 22 | PLAG1 | 1421745_at 1443943_at | 7126 | 0.176 | -0.0915 | No | ||
| 23 | ZFPM1 | 1423603_at 1451046_at | 7254 | 0.170 | -0.0934 | No | ||
| 24 | SPINK4 | 1427119_at | 8313 | 0.129 | -0.1389 | No | ||
| 25 | PRG4 | 1449824_at | 8421 | 0.125 | -0.1410 | No | ||
| 26 | CREB5 | 1440089_at 1442576_at 1457222_at | 8760 | 0.113 | -0.1539 | No | ||
| 27 | KYNU | 1430570_at 1451903_at | 8815 | 0.111 | -0.1538 | No | ||
| 28 | LRRN1 | 1416053_at 1432715_at 1432716_at | 8905 | 0.108 | -0.1555 | No | ||
| 29 | KCNJ16 | 1435094_at 1436163_at | 9139 | 0.100 | -0.1639 | No | ||
| 30 | MAP4K5 | 1427083_a_at 1427084_a_at 1427376_a_at 1440059_at 1445426_at 1446348_at 1447204_at | 9378 | 0.091 | -0.1727 | No | ||
| 31 | RASD1 | 1423619_at | 9400 | 0.090 | -0.1716 | No | ||
| 32 | ZNF76 | 1434085_at 1442446_at | 9716 | 0.080 | -0.1842 | No | ||
| 33 | KLF14 | 1457397_at | 9780 | 0.078 | -0.1853 | No | ||
| 34 | BMP10 | 1421763_at | 9899 | 0.074 | -0.1890 | No | ||
| 35 | TSHB | 1450371_at | 10183 | 0.064 | -0.2005 | No | ||
| 36 | GPR23 | 1434186_at 1439665_at 1452424_at | 10874 | 0.041 | -0.2311 | No | ||
| 37 | PTGFR | 1420349_at 1440777_x_at 1446331_at 1449828_at 1453924_a_at | 11271 | 0.028 | -0.2486 | No | ||
| 38 | GCG | 1425952_a_at | 11327 | 0.026 | -0.2506 | No | ||
| 39 | ASB2 | 1428444_at 1436259_at | 11713 | 0.013 | -0.2679 | No | ||
| 40 | PDE7A | 1423313_at 1423314_s_at 1450933_at 1451839_a_at 1458218_s_at | 11867 | 0.008 | -0.2747 | No | ||
| 41 | NRP1 | 1418084_at 1448943_at 1448944_at 1457198_at | 12136 | -0.001 | -0.2870 | No | ||
| 42 | RAB11FIP1 | 1430594_at 1447264_at | 12458 | -0.014 | -0.3013 | No | ||
| 43 | OTX2 | 1425926_a_at | 12558 | -0.018 | -0.3055 | No | ||
| 44 | ZFPM2 | 1419012_at 1444803_at 1447020_at 1449314_at | 12608 | -0.020 | -0.3073 | No | ||
| 45 | GPR44 | 1422275_at | 12849 | -0.028 | -0.3176 | No | ||
| 46 | NRAS | 1422688_a_at 1454060_a_at | 12913 | -0.030 | -0.3198 | No | ||
| 47 | IL7 | 1422080_at 1436861_at | 13120 | -0.038 | -0.3284 | No | ||
| 48 | TITF1 | 1422346_at | 13256 | -0.043 | -0.3336 | No | ||
| 49 | ACCN1 | 1417994_a_at | 13520 | -0.054 | -0.3444 | No | ||
| 50 | RASGRF2 | 1421621_at | 13793 | -0.064 | -0.3554 | No | ||
| 51 | CASQ2 | 1422529_s_at | 13835 | -0.066 | -0.3558 | No | ||
| 52 | RAI1 | 1420454_at 1453200_at | 13885 | -0.068 | -0.3565 | No | ||
| 53 | MYCT1 | 1426433_at 1438048_at 1447584_s_at 1452072_at | 13982 | -0.072 | -0.3592 | No | ||
| 54 | NDST2 | 1417931_at | 13994 | -0.072 | -0.3581 | No | ||
| 55 | CHCHD7 | 1440158_x_at 1444318_at 1446844_at 1454640_at | 14179 | -0.080 | -0.3647 | No | ||
| 56 | FHL3 | 1420779_at | 14401 | -0.090 | -0.3728 | No | ||
| 57 | HOXA1 | 1420565_at | 14533 | -0.096 | -0.3766 | No | ||
| 58 | HOXD10 | 1418606_at | 14688 | -0.103 | -0.3813 | No | ||
| 59 | PROX1 | 1421336_at 1437894_at 1457432_at | 15259 | -0.129 | -0.4044 | No | ||
| 60 | MYL6 | 1435041_at | 15368 | -0.134 | -0.4063 | No | ||
| 61 | UBXD3 | 1426007_a_at 1451906_at 1456385_x_at 1457581_at | 15491 | -0.141 | -0.4087 | No | ||
| 62 | IL4 | 1449864_at | 15504 | -0.141 | -0.4060 | No | ||
| 63 | EVX1 | 1421627_at | 15545 | -0.144 | -0.4046 | No | ||
| 64 | ISL1 | 1422720_at 1450723_at | 16188 | -0.182 | -0.4298 | No | ||
| 65 | RHOG | 1422572_at | 16665 | -0.214 | -0.4468 | No | ||
| 66 | ADNP | 1423069_at | 16939 | -0.234 | -0.4540 | No | ||
| 67 | IFRG15 | 1418115_s_at 1418116_at 1448958_at 1458364_s_at | 17280 | -0.263 | -0.4635 | No | ||
| 68 | S100A16 | 1425560_a_at 1447676_x_at | 17354 | -0.269 | -0.4608 | No | ||
| 69 | LMO2 | 1454086_a_at | 17874 | -0.324 | -0.4771 | No | ||
| 70 | FBXO18 | 1452153_at | 18095 | -0.351 | -0.4792 | No | ||
| 71 | SLC25A14 | 1417154_at | 18473 | -0.410 | -0.4872 | Yes | ||
| 72 | ABCB7 | 1419931_at 1427490_at 1435006_s_at | 18664 | -0.442 | -0.4859 | Yes | ||
| 73 | CRAT | 1417008_at 1441919_x_at 1448544_at | 18692 | -0.446 | -0.4769 | Yes | ||
| 74 | SLC37A4 | 1417042_at | 18911 | -0.490 | -0.4758 | Yes | ||
| 75 | BCL11B | 1440137_at 1441762_at 1450339_a_at | 18928 | -0.493 | -0.4653 | Yes | ||
| 76 | WDR23 | 1448418_s_at 1460189_at | 19407 | -0.592 | -0.4737 | Yes | ||
| 77 | CPNE1 | 1433439_at | 19849 | -0.712 | -0.4777 | Yes | ||
| 78 | VAMP1 | 1421862_a_at 1421863_at | 20173 | -0.825 | -0.4738 | Yes | ||
| 79 | PKLR | 1421258_a_at 1421259_at 1438711_at | 20195 | -0.834 | -0.4558 | Yes | ||
| 80 | PTPRC | 1422124_a_at 1440165_at | 20336 | -0.899 | -0.4418 | Yes | ||
| 81 | ESRRG | 1421747_at 1455267_at 1457896_at | 20530 | -0.993 | -0.4280 | Yes | ||
| 82 | MBNL2 | 1433754_at 1436858_at 1446475_at 1455827_at 1457237_at 1457477_at | 20536 | -0.996 | -0.4056 | Yes | ||
| 83 | SLC9A9 | 1433719_at 1437488_at 1447149_at | 20608 | -1.044 | -0.3851 | Yes | ||
| 84 | DMD | 1417307_at 1430320_at 1446156_at 1448665_at 1457022_at | 20709 | -1.130 | -0.3640 | Yes | ||
| 85 | CNP | 1418980_a_at 1437341_x_at 1449296_a_at | 20822 | -1.206 | -0.3418 | Yes | ||
| 86 | TOB1 | 1423176_at 1440844_at | 21309 | -1.696 | -0.3255 | Yes | ||
| 87 | SRPK2 | 1417134_at 1417135_at 1417136_s_at 1431372_at 1435746_at 1440080_at 1442241_at 1442980_at 1447755_at 1448603_at | 21433 | -1.928 | -0.2873 | Yes | ||
| 88 | PPIL5 | 1427506_at 1436922_at 1452458_s_at | 21655 | -2.428 | -0.2422 | Yes | ||
| 89 | HIP2 | 1417186_at 1417187_at 1417188_s_at 1441012_at 1442671_at 1443148_at 1444212_at | 21737 | -2.749 | -0.1834 | Yes | ||
| 90 | BCL6 | 1421818_at 1450381_a_at | 21792 | -3.059 | -0.1164 | Yes | ||
| 91 | ARPC2 | 1437148_at 1442295_at | 21917 | -5.403 | 0.0008 | Yes |

