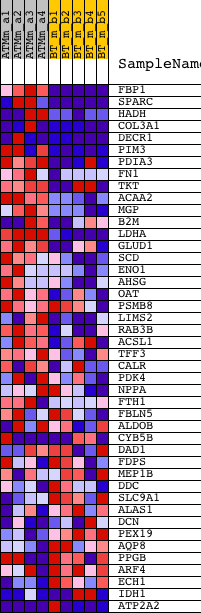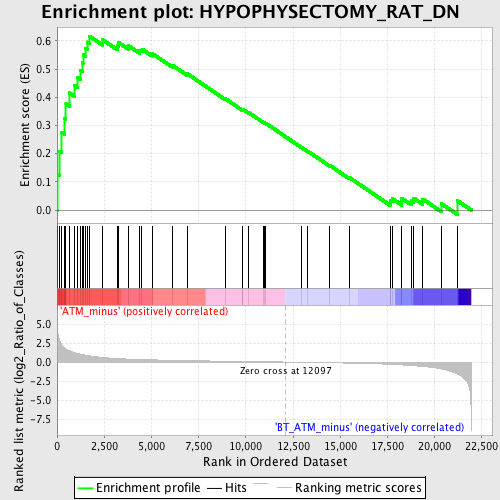
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | HYPOPHYSECTOMY_RAT_DN |
| Enrichment Score (ES) | 0.6165818 |
| Normalized Enrichment Score (NES) | 1.8022951 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.09449579 |
| FWER p-Value | 0.523 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | FBP1 | 1448470_at | 34 | 4.016 | 0.1260 | Yes | ||
| 2 | SPARC | 1416589_at 1448392_at 1458204_at | 146 | 2.707 | 0.2069 | Yes | ||
| 3 | HADH | 1436756_x_at 1455972_x_at 1460184_at | 226 | 2.295 | 0.2762 | Yes | ||
| 4 | COL3A1 | 1427883_a_at 1427884_at 1442977_at | 411 | 1.797 | 0.3248 | Yes | ||
| 5 | DECR1 | 1419367_at 1449443_at | 468 | 1.707 | 0.3765 | Yes | ||
| 6 | PIM3 | 1437100_x_at 1438956_x_at 1451069_at | 652 | 1.486 | 0.4153 | Yes | ||
| 7 | PDIA3 | 1423423_at | 931 | 1.228 | 0.4416 | Yes | ||
| 8 | FN1 | 1426642_at 1437218_at | 1083 | 1.121 | 0.4703 | Yes | ||
| 9 | TKT | 1436605_at 1439443_x_at 1451015_at | 1249 | 1.025 | 0.4953 | Yes | ||
| 10 | ACAA2 | 1428145_at 1428146_s_at 1455061_a_at | 1331 | 0.971 | 0.5225 | Yes | ||
| 11 | MGP | 1448416_at | 1373 | 0.951 | 0.5508 | Yes | ||
| 12 | B2M | 1427511_at 1449289_a_at 1452428_a_at | 1512 | 0.890 | 0.5728 | Yes | ||
| 13 | LDHA | 1419737_a_at | 1603 | 0.854 | 0.5958 | Yes | ||
| 14 | GLUD1 | 1416209_at 1448253_at | 1710 | 0.807 | 0.6166 | Yes | ||
| 15 | SCD | 1415964_at 1415965_at | 2414 | 0.601 | 0.6036 | No | ||
| 16 | ENO1 | 1419023_x_at | 3198 | 0.457 | 0.5823 | No | ||
| 17 | AHSG | 1455093_a_at | 3254 | 0.448 | 0.5940 | No | ||
| 18 | OAT | 1416452_at | 3767 | 0.386 | 0.5829 | No | ||
| 19 | PSMB8 | 1422962_a_at 1444619_x_at | 4378 | 0.333 | 0.5656 | No | ||
| 20 | LIMS2 | 1424408_at | 4493 | 0.325 | 0.5707 | No | ||
| 21 | RAB3B | 1422583_at | 5035 | 0.289 | 0.5552 | No | ||
| 22 | ACSL1 | 1422526_at 1423883_at 1447355_at 1450643_s_at 1460316_at | 6122 | 0.223 | 0.5126 | No | ||
| 23 | TFF3 | 1417370_at | 6905 | 0.185 | 0.4828 | No | ||
| 24 | CALR | 1417606_a_at 1433806_x_at 1456170_x_at | 8894 | 0.108 | 0.3954 | No | ||
| 25 | PDK4 | 1417273_at | 9810 | 0.077 | 0.3561 | No | ||
| 26 | NPPA | 1456062_at | 9843 | 0.076 | 0.3571 | No | ||
| 27 | FTH1 | 1448771_a_at | 10151 | 0.066 | 0.3451 | No | ||
| 28 | FBLN5 | 1416164_at 1446377_at | 10911 | 0.040 | 0.3117 | No | ||
| 29 | ALDOB | 1451194_at | 11002 | 0.037 | 0.3088 | No | ||
| 30 | CYB5B | 1417766_at 1417767_at 1443202_at 1448844_at | 11050 | 0.035 | 0.3077 | No | ||
| 31 | DAD1 | 1418528_a_at 1454860_x_at | 12966 | -0.033 | 0.2213 | No | ||
| 32 | FDPS | 1423418_at | 13276 | -0.043 | 0.2086 | No | ||
| 33 | MEP1B | 1418215_at | 14431 | -0.091 | 0.1587 | No | ||
| 34 | DDC | 1426215_at 1430591_at 1445524_at | 15476 | -0.140 | 0.1155 | No | ||
| 35 | SLC9A1 | 1417397_at | 17632 | -0.297 | 0.0265 | No | ||
| 36 | ALAS1 | 1424126_at 1442331_at 1455282_x_at | 17671 | -0.301 | 0.0343 | No | ||
| 37 | DCN | 1441506_at 1449368_at | 17747 | -0.310 | 0.0407 | No | ||
| 38 | PEX19 | 1416425_at 1442815_at 1448332_at 1455208_at | 18252 | -0.374 | 0.0296 | No | ||
| 39 | AQP8 | 1417828_at | 18259 | -0.374 | 0.0412 | No | ||
| 40 | PPGB | 1445659_at 1448128_at 1458466_at | 18756 | -0.458 | 0.0331 | No | ||
| 41 | ARF4 | 1423052_at 1423053_at 1434074_x_at 1439367_x_at 1442905_at 1457348_at | 18883 | -0.483 | 0.0427 | No | ||
| 42 | ECH1 | 1448491_at | 19362 | -0.583 | 0.0393 | No | ||
| 43 | IDH1 | 1419821_s_at 1422433_s_at | 20334 | -0.899 | 0.0235 | No | ||
| 44 | ATP2A2 | 1416551_at 1427250_at 1427251_at 1437797_at 1443551_at 1452363_a_at 1458927_at | 21190 | -1.559 | 0.0340 | No |