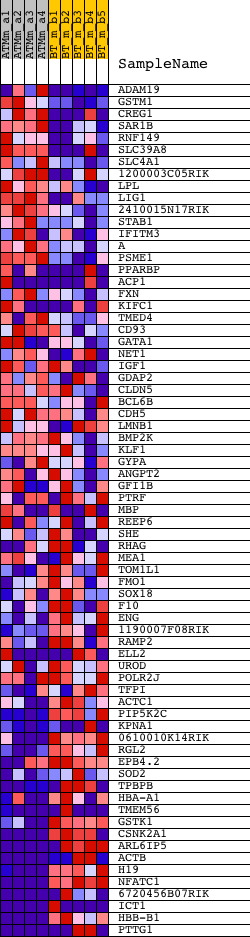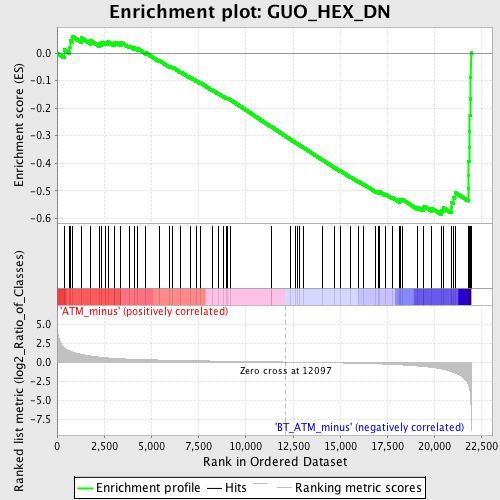
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | GUO_HEX_DN |
| Enrichment Score (ES) | -0.5862626 |
| Normalized Enrichment Score (NES) | -1.7887563 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.2679216 |
| FWER p-Value | 0.573 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ADAM19 | 1418402_at 1418403_at | 379 | 1.845 | 0.0122 | No | ||
| 2 | GSTM1 | 1416416_x_at 1448330_at | 673 | 1.470 | 0.0223 | No | ||
| 3 | CREG1 | 1415947_at 1415948_at | 685 | 1.457 | 0.0451 | No | ||
| 4 | SAR1B | 1428163_at | 791 | 1.353 | 0.0619 | No | ||
| 5 | RNF149 | 1429321_at | 1278 | 1.008 | 0.0558 | No | ||
| 6 | SLC39A8 | 1416832_at 1448482_at | 1786 | 0.783 | 0.0451 | No | ||
| 7 | SLC4A1 | 1416464_at 1431743_a_at 1434502_x_at | 2232 | 0.644 | 0.0350 | No | ||
| 8 | 1200003C05RIK | 1424106_at 1441959_s_at 1451238_at | 2365 | 0.613 | 0.0388 | No | ||
| 9 | LPL | 1415904_at | 2554 | 0.567 | 0.0393 | No | ||
| 10 | LIG1 | 1416641_at | 2697 | 0.535 | 0.0413 | No | ||
| 11 | 2410015N17RIK | 1416439_at | 3028 | 0.480 | 0.0339 | No | ||
| 12 | STAB1 | 1450199_a_at | 3052 | 0.476 | 0.0405 | No | ||
| 13 | IFITM3 | 1423754_at | 3353 | 0.435 | 0.0337 | No | ||
| 14 | A | 1420516_at 1455274_at 1460582_x_at | 3358 | 0.434 | 0.0404 | No | ||
| 15 | PSME1 | 1417056_at | 3815 | 0.381 | 0.0257 | No | ||
| 16 | PPARBP | 1421906_at 1421907_at 1439408_a_at 1446250_at 1447450_at 1448708_at 1450402_at 1453972_x_at | 4074 | 0.359 | 0.0196 | No | ||
| 17 | ACP1 | 1422716_a_at 1422717_at 1450720_at 1450721_at | 4272 | 0.341 | 0.0161 | No | ||
| 18 | FXN | 1427282_a_at | 4699 | 0.310 | 0.0015 | No | ||
| 19 | KIFC1 | 1449877_s_at 1456136_at | 5407 | 0.265 | -0.0266 | No | ||
| 20 | TMED4 | 1448422_at 1448423_at | 5972 | 0.231 | -0.0487 | No | ||
| 21 | CD93 | 1419589_at 1441907_s_at 1441908_x_at 1449521_at 1456046_at | 6092 | 0.225 | -0.0505 | No | ||
| 22 | GATA1 | 1449232_at | 6555 | 0.202 | -0.0684 | No | ||
| 23 | NET1 | 1421321_a_at 1457830_at | 7037 | 0.180 | -0.0875 | No | ||
| 24 | IGF1 | 1419519_at 1436497_at 1437401_at 1452014_a_at | 7375 | 0.165 | -0.1003 | No | ||
| 25 | GDAP2 | 1424057_at | 7605 | 0.154 | -0.1083 | No | ||
| 26 | CLDN5 | 1417839_at | 8223 | 0.132 | -0.1344 | No | ||
| 27 | BCL6B | 1418421_at | 8536 | 0.121 | -0.1468 | No | ||
| 28 | CDH5 | 1422047_at 1433956_at | 8819 | 0.111 | -0.1579 | No | ||
| 29 | LMNB1 | 1423520_at 1423521_at | 8986 | 0.105 | -0.1638 | No | ||
| 30 | BMP2K | 1421103_at 1437419_at 1458370_at | 9023 | 0.103 | -0.1638 | No | ||
| 31 | KLF1 | 1418600_at | 9161 | 0.099 | -0.1685 | No | ||
| 32 | GYPA | 1423016_a_at 1425643_at | 11336 | 0.026 | -0.2675 | No | ||
| 33 | ANGPT2 | 1422415_at 1446903_at 1448831_at | 11351 | 0.026 | -0.2678 | No | ||
| 34 | GFI1B | 1420399_at | 12340 | -0.009 | -0.3128 | No | ||
| 35 | PTRF | 1421431_at 1424130_a_at 1453891_at | 12601 | -0.020 | -0.3244 | No | ||
| 36 | MBP | 1419646_a_at 1425263_a_at 1425264_s_at 1433532_a_at 1436201_x_at 1451961_a_at 1454651_x_at 1456228_x_at | 12706 | -0.023 | -0.3288 | No | ||
| 37 | REEP6 | 1430128_a_at | 12857 | -0.029 | -0.3352 | No | ||
| 38 | SHE | 1438227_at 1457094_at | 13047 | -0.035 | -0.3432 | No | ||
| 39 | RHAG | 1419014_at | 14060 | -0.075 | -0.3883 | No | ||
| 40 | MEA1 | 1416810_at 1437943_s_at | 14705 | -0.104 | -0.4161 | No | ||
| 41 | TOM1L1 | 1439337_at 1451117_a_at | 15003 | -0.117 | -0.4278 | No | ||
| 42 | FMO1 | 1417429_at | 15546 | -0.144 | -0.4503 | No | ||
| 43 | SOX18 | 1449135_at | 15983 | -0.169 | -0.4676 | No | ||
| 44 | F10 | 1418992_at 1418993_s_at 1449305_at | 16251 | -0.185 | -0.4768 | No | ||
| 45 | ENG | 1417271_a_at 1432176_a_at | 16876 | -0.230 | -0.5017 | No | ||
| 46 | 1190007F08RIK | 1456641_at | 17044 | -0.243 | -0.5054 | No | ||
| 47 | RAMP2 | 1418187_at | 17094 | -0.248 | -0.5037 | No | ||
| 48 | ELL2 | 1450744_at 1458569_at | 17375 | -0.271 | -0.5122 | No | ||
| 49 | UROD | 1417206_at 1443849_x_at | 17744 | -0.309 | -0.5241 | No | ||
| 50 | POLR2J | 1417720_at 1440065_at | 18123 | -0.355 | -0.5357 | No | ||
| 51 | TFPI | 1438530_at 1451790_a_at 1451791_at 1452432_at | 18169 | -0.361 | -0.5320 | No | ||
| 52 | ACTC1 | 1415927_at | 18282 | -0.378 | -0.5311 | No | ||
| 53 | PIP5K2C | 1416387_at 1416388_at | 19090 | -0.525 | -0.5596 | No | ||
| 54 | KPNA1 | 1419548_at 1449503_at 1449504_at 1460260_s_at | 19391 | -0.587 | -0.5639 | No | ||
| 55 | 0610010K14RIK | 1428679_s_at 1429252_at | 19428 | -0.596 | -0.5560 | No | ||
| 56 | RGL2 | 1450688_at | 19847 | -0.712 | -0.5638 | No | ||
| 57 | EPB4.2 | 1417337_at 1417338_at | 20340 | -0.900 | -0.5719 | Yes | ||
| 58 | SOD2 | 1417193_at 1417194_at 1442994_at 1444531_at 1448610_a_at 1454976_at | 20453 | -0.952 | -0.5618 | Yes | ||
| 59 | TPBPB | 1416169_at 1456246_x_at | 20870 | -1.237 | -0.5610 | Yes | ||
| 60 | HBA-A1 | 1417714_x_at 1428361_x_at 1452757_s_at 1453574_at | 20881 | -1.246 | -0.5416 | Yes | ||
| 61 | TMEM56 | 1431404_at 1434553_at 1439353_x_at 1445744_at 1456718_at | 21004 | -1.361 | -0.5254 | Yes | ||
| 62 | GSTK1 | 1452823_at | 21073 | -1.426 | -0.5057 | Yes | ||
| 63 | CSNK2A1 | 1419034_at 1419035_s_at 1419036_at 1419037_at 1419038_a_at 1453427_at | 21764 | -2.889 | -0.4911 | Yes | ||
| 64 | ARL6IP5 | 1417225_at 1440360_at | 21796 | -3.100 | -0.4429 | Yes | ||
| 65 | ACTB | 1419734_at 1436722_a_at 1440365_at AFFX-b-ActinMur/M12481_3_at AFFX-b-ActinMur/M12481_5_at AFFX-b-ActinMur/M12481_M_at | 21800 | -3.133 | -0.3930 | Yes | ||
| 66 | H19 | 1448194_a_at | 21820 | -3.263 | -0.3417 | Yes | ||
| 67 | NFATC1 | 1417621_at 1425761_a_at 1428479_at 1447084_at 1447085_s_at | 21857 | -3.609 | -0.2856 | Yes | ||
| 68 | 6720456B07RIK | 1423188_a_at 1423189_at 1444806_at 1446299_at | 21864 | -3.682 | -0.2271 | Yes | ||
| 69 | ICT1 | 1444276_at 1460308_a_at | 21880 | -3.874 | -0.1658 | Yes | ||
| 70 | HBB-B1 | 1427866_x_at | 21907 | -4.893 | -0.0888 | Yes | ||
| 71 | PTTG1 | 1419620_at 1424105_a_at 1438390_s_at | 21919 | -5.628 | 0.0007 | Yes |