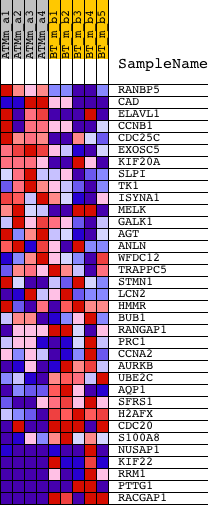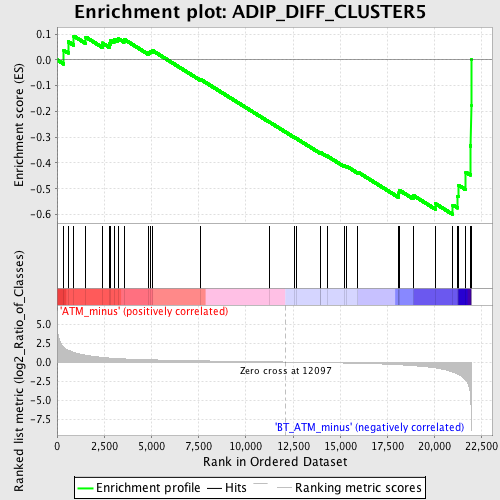
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | BT_ATM_minus |
| GeneSet | ADIP_DIFF_CLUSTER5 |
| Enrichment Score (ES) | -0.5999739 |
| Normalized Enrichment Score (NES) | -1.6323786 |
| Nominal p-value | 0.003629764 |
| FDR q-value | 0.34920123 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RANBP5 | 1426945_at 1426946_at 1431706_at | 356 | 1.898 | 0.0371 | No | ||
| 2 | CAD | 1440857_at 1452829_at 1452830_s_at | 590 | 1.555 | 0.0702 | No | ||
| 3 | ELAVL1 | 1428653_x_at 1431037_a_at 1440373_at 1440464_at 1445521_at 1447109_at 1448151_at 1452858_at | 887 | 1.263 | 0.0922 | No | ||
| 4 | CCNB1 | 1419943_s_at 1419944_at | 1521 | 0.886 | 0.0882 | No | ||
| 5 | CDC25C | 1422252_a_at | 2391 | 0.607 | 0.0656 | No | ||
| 6 | EXOSC5 | 1417167_at | 2775 | 0.523 | 0.0629 | No | ||
| 7 | KIF20A | 1449207_a_at | 2824 | 0.513 | 0.0751 | No | ||
| 8 | SLPI | 1448377_at | 3015 | 0.482 | 0.0800 | No | ||
| 9 | TK1 | 1416258_at | 3224 | 0.452 | 0.0832 | No | ||
| 10 | ISYNA1 | 1415977_at | 3544 | 0.410 | 0.0802 | No | ||
| 11 | MELK | 1416558_at | 4828 | 0.302 | 0.0301 | No | ||
| 12 | GALK1 | 1417177_at | 4921 | 0.296 | 0.0342 | No | ||
| 13 | AGT | 1423396_at | 5070 | 0.287 | 0.0355 | No | ||
| 14 | ANLN | 1433543_at 1439648_at | 7583 | 0.155 | -0.0748 | No | ||
| 15 | WFDC12 | 1449191_at | 11262 | 0.028 | -0.2420 | No | ||
| 16 | TRAPPC5 | 1418233_a_at 1448999_at | 12570 | -0.019 | -0.3012 | No | ||
| 17 | STMN1 | 1415849_s_at | 12666 | -0.021 | -0.3049 | No | ||
| 18 | LCN2 | 1427747_a_at | 13948 | -0.070 | -0.3614 | No | ||
| 19 | HMMR | 1425815_a_at 1427541_x_at 1429871_at 1450156_a_at 1450157_a_at | 13951 | -0.070 | -0.3595 | No | ||
| 20 | BUB1 | 1424046_at 1438571_at | 14295 | -0.085 | -0.3728 | No | ||
| 21 | RANGAP1 | 1423749_s_at 1444581_at 1451092_a_at | 15201 | -0.127 | -0.4105 | No | ||
| 22 | PRC1 | 1423774_a_at 1423775_s_at | 15319 | -0.132 | -0.4122 | No | ||
| 23 | CCNA2 | 1417910_at 1417911_at | 15928 | -0.166 | -0.4353 | No | ||
| 24 | AURKB | 1424128_x_at 1451246_s_at | 18073 | -0.348 | -0.5234 | No | ||
| 25 | UBE2C | 1452954_at | 18103 | -0.351 | -0.5148 | No | ||
| 26 | AQP1 | 1416203_at | 18113 | -0.353 | -0.5053 | No | ||
| 27 | SFRS1 | 1428099_a_at 1428100_at 1430982_at 1434972_x_at 1452430_s_at 1453722_s_at 1457136_at | 18856 | -0.477 | -0.5258 | No | ||
| 28 | H2AFX | 1416746_at | 20032 | -0.771 | -0.5577 | No | ||
| 29 | CDC20 | 1416664_at 1439377_x_at 1439394_x_at | 20958 | -1.309 | -0.5632 | Yes | ||
| 30 | S100A8 | 1419394_s_at | 21216 | -1.589 | -0.5302 | Yes | ||
| 31 | NUSAP1 | 1416309_at 1416310_at 1442830_at | 21255 | -1.634 | -0.4860 | Yes | ||
| 32 | KIF22 | 1423813_at 1437716_x_at 1451128_s_at | 21648 | -2.409 | -0.4362 | Yes | ||
| 33 | RRM1 | 1415878_at 1440073_at 1448127_at | 21885 | -4.048 | -0.3331 | Yes | ||
| 34 | PTTG1 | 1419620_at 1424105_a_at 1438390_s_at | 21919 | -5.628 | -0.1763 | Yes | ||
| 35 | RACGAP1 | 1421546_a_at 1441889_x_at 1451358_a_at | 21926 | -6.293 | 0.0004 | Yes |