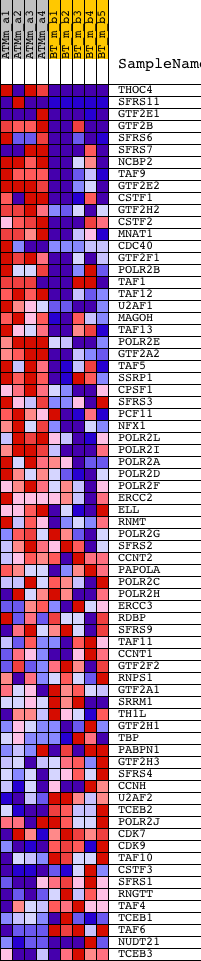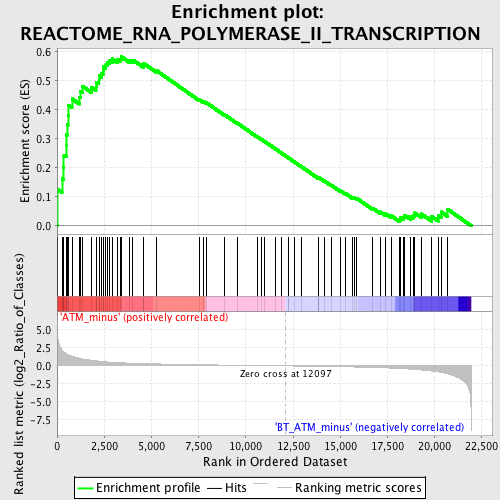
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus.phenotype_ATM_minus_versus_BT_ATM_minus.cls #ATM_minus_versus_BT_ATM_minus_repos |
| Phenotype | phenotype_ATM_minus_versus_BT_ATM_minus.cls#ATM_minus_versus_BT_ATM_minus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | REACTOME_RNA_POLYMERASE_II_TRANSCRIPTION |
| Enrichment Score (ES) | 0.58244306 |
| Normalized Enrichment Score (NES) | 1.8077893 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.030582853 |
| FWER p-Value | 0.289 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | THOC4 | 1417724_at 1436211_at | 7 | 5.638 | 0.1263 | Yes | ||
| 2 | SFRS11 | 1427269_at 1430077_at 1439095_at 1452371_at | 260 | 2.144 | 0.1630 | Yes | ||
| 3 | GTF2E1 | 1428124_at 1443110_at | 339 | 1.924 | 0.2026 | Yes | ||
| 4 | GTF2B | 1438066_at 1451135_at | 362 | 1.878 | 0.2438 | Yes | ||
| 5 | SFRS6 | 1416720_at 1416721_s_at 1447898_s_at 1448454_at | 485 | 1.692 | 0.2762 | Yes | ||
| 6 | SFRS7 | 1424033_at 1424883_s_at 1436871_at | 488 | 1.687 | 0.3140 | Yes | ||
| 7 | NCBP2 | 1423045_at 1423046_s_at 1450847_at | 527 | 1.641 | 0.3491 | Yes | ||
| 8 | TAF9 | 1422778_at 1451509_at | 578 | 1.566 | 0.3820 | Yes | ||
| 9 | GTF2E2 | 1448884_at | 593 | 1.550 | 0.4162 | Yes | ||
| 10 | CSTF1 | 1417117_at 1448597_at | 790 | 1.354 | 0.4376 | Yes | ||
| 11 | GTF2H2 | 1450701_a_at 1457818_at | 1179 | 1.069 | 0.4439 | Yes | ||
| 12 | CSTF2 | 1419644_at 1419645_at 1455523_at | 1239 | 1.033 | 0.4644 | Yes | ||
| 13 | MNAT1 | 1432177_a_at | 1351 | 0.962 | 0.4809 | Yes | ||
| 14 | CDC40 | 1442667_at 1445348_at 1453342_at 1457366_at | 1823 | 0.769 | 0.4767 | Yes | ||
| 15 | GTF2F1 | 1417698_at 1417699_at | 2060 | 0.694 | 0.4815 | Yes | ||
| 16 | POLR2B | 1433552_a_at | 2086 | 0.687 | 0.4957 | Yes | ||
| 17 | TAF1 | 1447417_at 1455513_at 1458744_at | 2218 | 0.651 | 0.5044 | Yes | ||
| 18 | TAF12 | 1423398_at | 2229 | 0.645 | 0.5184 | Yes | ||
| 19 | U2AF1 | 1422509_at | 2374 | 0.611 | 0.5255 | Yes | ||
| 20 | MAGOH | 1416212_at | 2436 | 0.596 | 0.5361 | Yes | ||
| 21 | TAF13 | 1448707_at | 2455 | 0.590 | 0.5486 | Yes | ||
| 22 | POLR2E | 1417138_s_at 1451093_at 1458326_at | 2587 | 0.560 | 0.5552 | Yes | ||
| 23 | GTF2A2 | 1433528_at | 2647 | 0.544 | 0.5647 | Yes | ||
| 24 | TAF5 | 1435474_at | 2768 | 0.524 | 0.5710 | Yes | ||
| 25 | SSRP1 | 1426788_a_at 1426789_s_at 1426790_at | 2915 | 0.499 | 0.5755 | Yes | ||
| 26 | CPSF1 | 1417665_a_at | 3195 | 0.457 | 0.5730 | Yes | ||
| 27 | SFRS3 | 1416150_a_at 1416151_at 1416152_a_at 1454993_a_at | 3375 | 0.432 | 0.5745 | Yes | ||
| 28 | PCF11 | 1427159_at 1427160_at 1428724_at 1456489_at | 3413 | 0.428 | 0.5824 | Yes | ||
| 29 | NFX1 | 1419752_at 1419753_at 1428248_at 1442397_at | 3847 | 0.377 | 0.5711 | No | ||
| 30 | POLR2L | 1428296_at | 4017 | 0.364 | 0.5715 | No | ||
| 31 | POLR2I | 1428494_a_at | 4579 | 0.319 | 0.5530 | No | ||
| 32 | POLR2A | 1422311_a_at 1426242_at 1458710_at | 4584 | 0.318 | 0.5600 | No | ||
| 33 | POLR2D | 1424258_at | 5247 | 0.275 | 0.5359 | No | ||
| 34 | POLR2F | 1415754_at 1427402_at | 7531 | 0.158 | 0.4350 | No | ||
| 35 | ERCC2 | 1460727_at | 7740 | 0.150 | 0.4289 | No | ||
| 36 | ELL | 1441928_x_at 1460643_at | 7901 | 0.144 | 0.4248 | No | ||
| 37 | RNMT | 1436328_at 1440002_at 1446442_at 1453106_a_at | 8854 | 0.110 | 0.3837 | No | ||
| 38 | POLR2G | 1416270_at | 9534 | 0.086 | 0.3546 | No | ||
| 39 | SFRS2 | 1415807_s_at 1427504_s_at 1427816_at 1452439_s_at | 10600 | 0.050 | 0.3070 | No | ||
| 40 | CCNT2 | 1427088_at 1427089_at 1435445_at 1452297_at 1453399_at 1457439_at | 10842 | 0.042 | 0.2970 | No | ||
| 41 | PAPOLA | 1424216_a_at 1424217_at 1427544_a_at 1444015_at 1455836_at | 10992 | 0.037 | 0.2910 | No | ||
| 42 | POLR2C | 1416341_at | 11571 | 0.018 | 0.2649 | No | ||
| 43 | POLR2H | 1424473_at | 11897 | 0.007 | 0.2502 | No | ||
| 44 | ERCC3 | 1448497_at | 12233 | -0.005 | 0.2350 | No | ||
| 45 | RDBP | 1450705_at | 12251 | -0.005 | 0.2343 | No | ||
| 46 | SFRS9 | 1417727_at | 12595 | -0.019 | 0.2191 | No | ||
| 47 | TAF11 | 1446161_at 1451995_at | 12926 | -0.031 | 0.2047 | No | ||
| 48 | CCNT1 | 1419313_at 1456477_at | 13828 | -0.066 | 0.1649 | No | ||
| 49 | GTF2F2 | 1426626_at 1443233_at | 13838 | -0.066 | 0.1660 | No | ||
| 50 | RNPS1 | 1437027_x_at 1437359_at 1437851_x_at 1438267_x_at 1448324_at | 13848 | -0.066 | 0.1671 | No | ||
| 51 | GTF2A1 | 1421357_at | 14157 | -0.080 | 0.1548 | No | ||
| 52 | SRRM1 | 1420934_a_at 1420935_a_at 1442330_at 1447447_s_at 1450045_at 1454689_at 1459828_at | 14513 | -0.095 | 0.1407 | No | ||
| 53 | TH1L | 1416198_at | 14981 | -0.116 | 0.1219 | No | ||
| 54 | GTF2H1 | 1453169_a_at | 15270 | -0.130 | 0.1117 | No | ||
| 55 | TBP | 1426469_a_at 1426470_at | 15631 | -0.149 | 0.0985 | No | ||
| 56 | PABPN1 | 1415728_at 1422848_a_at 1422849_a_at 1422850_at | 15749 | -0.156 | 0.0967 | No | ||
| 57 | GTF2H3 | 1454092_a_at | 15860 | -0.162 | 0.0953 | No | ||
| 58 | SFRS4 | 1448778_at | 16705 | -0.217 | 0.0616 | No | ||
| 59 | CCNH | 1418585_at | 17127 | -0.250 | 0.0479 | No | ||
| 60 | U2AF2 | 1417260_at | 17408 | -0.274 | 0.0413 | No | ||
| 61 | TCEB2 | 1428263_a_at | 17707 | -0.305 | 0.0345 | No | ||
| 62 | POLR2J | 1417720_at 1440065_at | 18123 | -0.355 | 0.0235 | No | ||
| 63 | CDK7 | 1424257_at 1439511_at 1451741_a_at 1458987_at | 18206 | -0.367 | 0.0280 | No | ||
| 64 | CDK9 | 1417269_at 1457240_at | 18362 | -0.391 | 0.0297 | No | ||
| 65 | TAF10 | 1448784_at | 18407 | -0.397 | 0.0366 | No | ||
| 66 | CSTF3 | 1424723_s_at 1443909_at | 18724 | -0.451 | 0.0322 | No | ||
| 67 | SFRS1 | 1428099_a_at 1428100_at 1430982_at 1434972_x_at 1452430_s_at 1453722_s_at 1457136_at | 18856 | -0.477 | 0.0370 | No | ||
| 68 | RNGTT | 1425844_a_at 1448714_at | 18936 | -0.495 | 0.0445 | No | ||
| 69 | TAF4 | 1427460_at 1452438_s_at | 19291 | -0.568 | 0.0411 | No | ||
| 70 | TCEB1 | 1427914_a_at 1427915_s_at 1436226_at 1452593_a_at | 19835 | -0.709 | 0.0322 | No | ||
| 71 | TAF6 | 1418593_at | 20174 | -0.825 | 0.0352 | No | ||
| 72 | NUDT21 | 1417681_at 1428339_at 1437213_at 1455966_s_at | 20339 | -0.900 | 0.0479 | No | ||
| 73 | TCEB3 | 1422591_at 1434117_at 1450676_at | 20682 | -1.111 | 0.0573 | No |