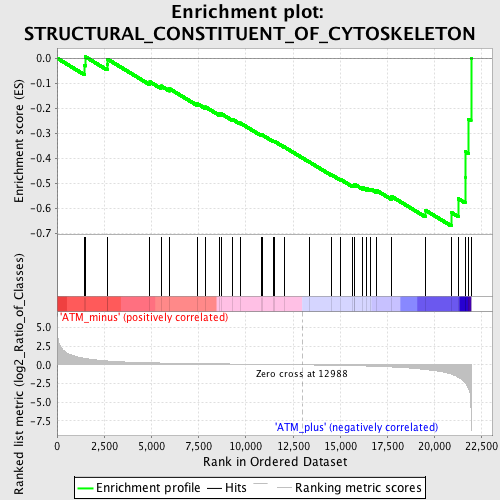
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | STRUCTURAL_CONSTITUENT_OF_CYTOSKELETON |
| Enrichment Score (ES) | -0.6696382 |
| Normalized Enrichment Score (NES) | -1.722217 |
| Nominal p-value | 0.0017921147 |
| FDR q-value | 0.78026754 |
| FWER p-Value | 0.782 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | LOR | 1420183_at 1448745_s_at | 1446 | 0.869 | -0.0286 | No | ||
| 2 | BFSP1 | 1450571_a_at | 1503 | 0.849 | 0.0053 | No | ||
| 3 | ADD3 | 1423297_at 1423298_at 1426574_a_at 1447077_at 1456162_x_at | 2652 | 0.514 | -0.0250 | No | ||
| 4 | VIM | 1438118_x_at 1450641_at 1456292_a_at | 2674 | 0.510 | -0.0040 | No | ||
| 5 | DSP | 1427610_at 1435493_at 1435494_s_at | 4917 | 0.275 | -0.0945 | No | ||
| 6 | ACTL6B | 1422564_at | 5503 | 0.243 | -0.1108 | No | ||
| 7 | VILL | 1426022_a_at | 5946 | 0.223 | -0.1214 | No | ||
| 8 | SORBS2 | 1437197_at 1441624_at 1446744_at | 7434 | 0.167 | -0.1821 | No | ||
| 9 | TUBG1 | 1417144_at | 7853 | 0.154 | -0.1946 | No | ||
| 10 | HIP1 | 1424755_at 1424756_at 1432017_at 1432230_at 1434557_at 1444873_at | 8580 | 0.131 | -0.2221 | No | ||
| 11 | SORBS3 | 1419329_at | 8690 | 0.127 | -0.2216 | No | ||
| 12 | ARPC1B | 1416226_at | 9267 | 0.109 | -0.2432 | No | ||
| 13 | DES | 1426731_at | 9694 | 0.097 | -0.2585 | No | ||
| 14 | INA | 1418178_at 1448991_a_at 1448992_at | 10812 | 0.064 | -0.3068 | No | ||
| 15 | ACTL7A | 1419009_at | 10858 | 0.063 | -0.3061 | No | ||
| 16 | MAPT | 1417885_at 1424718_at 1424719_a_at 1445634_at 1455028_at | 11464 | 0.046 | -0.3318 | No | ||
| 17 | NEFL | 1426255_at | 11502 | 0.045 | -0.3315 | No | ||
| 18 | GFAP | 1426508_at 1426509_s_at 1440142_s_at | 12015 | 0.029 | -0.3536 | No | ||
| 19 | DST | 1421117_at 1421276_a_at 1423626_at 1442395_at 1446943_at 1450119_at 1458075_at 1459098_at 1459356_at | 13356 | -0.013 | -0.4143 | No | ||
| 20 | ANK1 | 1419421_at 1425677_a_at 1443291_at 1450627_at 1452512_a_at | 14540 | -0.056 | -0.4659 | No | ||
| 21 | ACTL7B | 1424678_at | 15019 | -0.076 | -0.4844 | No | ||
| 22 | ARPC3 | 1448279_at | 15629 | -0.106 | -0.5076 | No | ||
| 23 | BFSP2 | 1434463_at 1459306_at | 15757 | -0.114 | -0.5085 | No | ||
| 24 | CD2AP | 1420906_at 1420907_at 1420908_at 1460140_at | 15765 | -0.114 | -0.5040 | No | ||
| 25 | PPL | 1460732_a_at | 16191 | -0.141 | -0.5173 | No | ||
| 26 | ARPC4 | 1423588_at 1423589_at | 16393 | -0.155 | -0.5198 | No | ||
| 27 | SPTBN4 | 1425116_a_at | 16609 | -0.171 | -0.5223 | No | ||
| 28 | BICD1 | 1427530_at 1437548_at 1438701_at 1451684_a_at | 16916 | -0.198 | -0.5277 | No | ||
| 29 | ARPC5 | 1444568_at 1448129_at | 17712 | -0.286 | -0.5517 | No | ||
| 30 | ACTA1 | 1427735_a_at | 19508 | -0.620 | -0.6070 | No | ||
| 31 | DMD | 1417307_at 1430320_at 1446156_at 1448665_at 1457022_at | 20881 | -1.246 | -0.6160 | Yes | ||
| 32 | MSN | 1421814_at 1450379_at | 21252 | -1.666 | -0.5613 | Yes | ||
| 33 | TUBE1 | 1431873_a_at 1437942_x_at 1443481_at | 21608 | -2.372 | -0.4754 | Yes | ||
| 34 | CCDC6 | 1428311_at 1459159_a_at | 21615 | -2.396 | -0.3727 | Yes | ||
| 35 | ACTB | 1419734_at 1436722_a_at 1440365_at AFFX-b-ActinMur/M12481_3_at AFFX-b-ActinMur/M12481_5_at AFFX-b-ActinMur/M12481_M_at | 21797 | -3.181 | -0.2440 | Yes | ||
| 36 | ARPC2 | 1437148_at 1442295_at | 21921 | -5.817 | 0.0006 | Yes |

