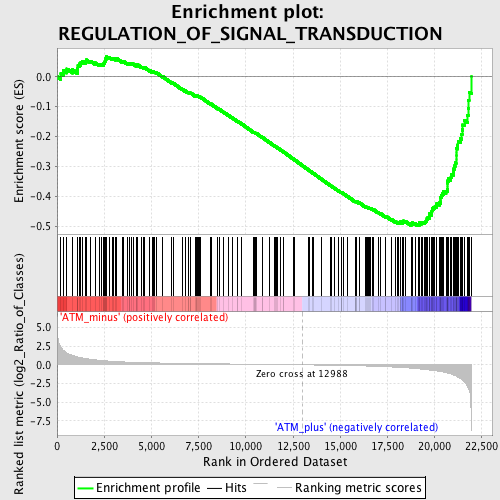
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | REGULATION_OF_SIGNAL_TRANSDUCTION |
| Enrichment Score (ES) | -0.4984058 |
| Normalized Enrichment Score (NES) | -1.6531311 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.35784173 |
| FWER p-Value | 0.984 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | VAPA | 1416465_a_at 1416466_at 1448349_at | 203 | 2.343 | 0.0107 | No | ||
| 2 | HMOX1 | 1448239_at | 321 | 1.931 | 0.0217 | No | ||
| 3 | UBE2N | 1422559_at 1435384_at | 522 | 1.575 | 0.0260 | No | ||
| 4 | TGFB1 | 1420653_at 1445360_at | 817 | 1.249 | 0.0231 | No | ||
| 5 | ECM1 | 1448613_at | 1084 | 1.067 | 0.0200 | No | ||
| 6 | TFG | 1415887_at 1438053_at | 1095 | 1.062 | 0.0286 | No | ||
| 7 | CITED2 | 1417129_a_at 1421267_a_at 1439915_at 1440091_at 1443561_at 1443926_at 1445638_at 1446176_at 1447861_x_at 1457632_s_at 1459301_at | 1105 | 1.052 | 0.0372 | No | ||
| 8 | PTPRC | 1422124_a_at 1440165_at | 1172 | 1.016 | 0.0428 | No | ||
| 9 | BST2 | 1424921_at | 1260 | 0.968 | 0.0471 | No | ||
| 10 | CBLC | 1422666_at 1431692_a_at | 1333 | 0.927 | 0.0517 | No | ||
| 11 | GNG7 | 1455190_at | 1528 | 0.839 | 0.0499 | No | ||
| 12 | ECT2 | 1419513_a_at | 1545 | 0.832 | 0.0562 | No | ||
| 13 | EEF1D | 1428135_a_at 1439439_x_at 1449506_a_at 1459754_x_at | 1763 | 0.746 | 0.0526 | No | ||
| 14 | LGALS9 | 1421217_a_at | 2026 | 0.660 | 0.0462 | No | ||
| 15 | RPL17 | 1423855_x_at 1435791_x_at 1453752_at | 2218 | 0.607 | 0.0426 | No | ||
| 16 | GPR89A | 1424696_at | 2359 | 0.578 | 0.0411 | No | ||
| 17 | GJA1 | 1415800_at 1415801_at 1437992_x_at 1438650_x_at 1438945_x_at 1438973_x_at | 2449 | 0.557 | 0.0418 | No | ||
| 18 | PPM1A | 1415678_at 1417221_at 1425537_at 1429500_at 1429501_s_at 1443903_at 1451943_a_at 1453171_s_at | 2476 | 0.550 | 0.0453 | No | ||
| 19 | PLK2 | 1427005_at | 2523 | 0.539 | 0.0478 | No | ||
| 20 | RIC8B | 1435096_at 1439724_at | 2534 | 0.538 | 0.0519 | No | ||
| 21 | CD24 | 1416034_at 1437502_x_at 1448182_a_at | 2551 | 0.535 | 0.0557 | No | ||
| 22 | ATP6AP2 | 1423662_at 1437688_x_at 1439456_x_at | 2564 | 0.532 | 0.0597 | No | ||
| 23 | GOLT1B | 1449859_at 1460583_at | 2593 | 0.525 | 0.0629 | No | ||
| 24 | TNFRSF10B | 1421296_at 1422344_s_at | 2612 | 0.522 | 0.0665 | No | ||
| 25 | MAPK8IP3 | 1416437_a_at 1425975_a_at 1436676_at 1460628_at | 2765 | 0.489 | 0.0637 | No | ||
| 26 | GRK1 | 1421361_at | 2919 | 0.462 | 0.0606 | No | ||
| 27 | TRAF3IP2 | 1445985_at 1448508_at | 2977 | 0.453 | 0.0619 | No | ||
| 28 | LTBR | 1416435_at | 3111 | 0.432 | 0.0594 | No | ||
| 29 | LGALS1 | 1419573_a_at 1455439_a_at | 3168 | 0.426 | 0.0605 | No | ||
| 30 | SHC1 | 1422853_at 1422854_at | 3459 | 0.392 | 0.0505 | No | ||
| 31 | SMAD2 | 1420634_a_at 1458304_at | 3526 | 0.385 | 0.0507 | No | ||
| 32 | SOST | 1421245_at 1436240_at 1450179_at | 3714 | 0.363 | 0.0452 | No | ||
| 33 | TSPAN6 | 1416872_at 1448501_at | 3825 | 0.352 | 0.0432 | No | ||
| 34 | GRM4 | 1457299_at | 3855 | 0.349 | 0.0448 | No | ||
| 35 | TPD52L1 | 1418412_at | 3939 | 0.342 | 0.0439 | No | ||
| 36 | TRIM13 | 1417888_at | 4027 | 0.334 | 0.0428 | No | ||
| 37 | GLI1 | 1449058_at | 4186 | 0.321 | 0.0382 | No | ||
| 38 | GRK4 | 1420355_at 1454152_a_at | 4220 | 0.320 | 0.0395 | No | ||
| 39 | RGS9 | 1418691_at 1426033_at 1439635_at | 4245 | 0.318 | 0.0411 | No | ||
| 40 | TDGF1 | 1450989_at | 4494 | 0.301 | 0.0322 | No | ||
| 41 | CASP8 | 1424552_at | 4574 | 0.296 | 0.0311 | No | ||
| 42 | ADRB3 | 1421555_at 1455918_at | 4652 | 0.291 | 0.0301 | No | ||
| 43 | ZDHHC17 | 1434397_at 1447656_at 1455986_at 1458084_at 1458363_at | 4902 | 0.276 | 0.0210 | No | ||
| 44 | PEG10 | 1427550_at | 5051 | 0.267 | 0.0164 | No | ||
| 45 | ABRA | 1458455_at | 5080 | 0.265 | 0.0174 | No | ||
| 46 | NFAM1 | 1425714_a_at 1428790_at | 5182 | 0.260 | 0.0150 | No | ||
| 47 | NF1 | 1438067_at 1441523_at 1443097_at 1452525_a_at | 5271 | 0.256 | 0.0131 | No | ||
| 48 | CDKN1C | 1417649_at | 5595 | 0.239 | 0.0003 | No | ||
| 49 | NEK6 | 1451043_at | 6069 | 0.218 | -0.0196 | No | ||
| 50 | NCK2 | 1416796_at 1416797_at 1458432_at | 6147 | 0.214 | -0.0213 | No | ||
| 51 | CIDEA | 1417956_at | 6634 | 0.195 | -0.0420 | No | ||
| 52 | RAC1 | 1423734_at 1437674_at 1451086_s_at | 6772 | 0.190 | -0.0467 | No | ||
| 53 | RGS5 | 1420940_x_at 1420941_at | 6935 | 0.185 | -0.0526 | No | ||
| 54 | FRZB | 1416658_at 1448424_at | 6975 | 0.183 | -0.0528 | No | ||
| 55 | NDFIP1 | 1424820_a_at 1424821_at 1451493_at | 7047 | 0.181 | -0.0545 | No | ||
| 56 | PPP5C | 1424115_at 1424116_x_at 1451242_a_at | 7071 | 0.180 | -0.0540 | No | ||
| 57 | CDC42BPA | 1435847_at 1436380_at 1438449_at 1442260_at 1446945_at | 7303 | 0.172 | -0.0632 | No | ||
| 58 | IGF1 | 1419519_at 1436497_at 1437401_at 1452014_a_at | 7365 | 0.169 | -0.0646 | No | ||
| 59 | LITAF | 1416303_at 1416304_at | 7383 | 0.169 | -0.0639 | No | ||
| 60 | RHOC | 1435394_s_at 1448605_at | 7414 | 0.168 | -0.0638 | No | ||
| 61 | PLEKHG5 | 1452248_at | 7465 | 0.167 | -0.0647 | No | ||
| 62 | IL12A | 1425454_a_at | 7539 | 0.164 | -0.0667 | No | ||
| 63 | GRK5 | 1446361_at 1446998_at 1449514_at | 7589 | 0.163 | -0.0675 | No | ||
| 64 | EREG | 1419431_at | 8099 | 0.145 | -0.0897 | No | ||
| 65 | ACVR2A | 1437382_at 1439643_at 1445026_at 1451004_at | 8150 | 0.143 | -0.0908 | No | ||
| 66 | TLR6 | 1421352_at | 8519 | 0.133 | -0.1066 | No | ||
| 67 | NF2 | 1421820_a_at 1427708_a_at 1450382_at 1451829_a_at | 8584 | 0.131 | -0.1084 | No | ||
| 68 | TMED4 | 1448422_at 1448423_at | 8813 | 0.123 | -0.1179 | No | ||
| 69 | CASP1 | 1449265_at | 9080 | 0.115 | -0.1291 | No | ||
| 70 | IL20 | 1421608_at | 9287 | 0.109 | -0.1377 | No | ||
| 71 | MFN2 | 1441424_at 1448131_at 1451900_at | 9543 | 0.101 | -0.1485 | No | ||
| 72 | AMBP | 1416649_at | 9577 | 0.100 | -0.1492 | No | ||
| 73 | SMAD3 | 1450471_at 1450472_s_at 1454960_at | 9779 | 0.094 | -0.1576 | No | ||
| 74 | FGD5 | 1460578_at | 10393 | 0.077 | -0.1852 | No | ||
| 75 | MAPK8IP2 | 1418785_at 1418786_at 1435045_s_at 1449225_a_at 1455194_at | 10445 | 0.075 | -0.1869 | No | ||
| 76 | SECTM1 | 1419478_at 1419479_at | 10461 | 0.075 | -0.1869 | No | ||
| 77 | TNFSF10 | 1420412_at 1439680_at 1459913_at | 10505 | 0.074 | -0.1883 | No | ||
| 78 | RGN | 1448852_at | 10518 | 0.073 | -0.1882 | No | ||
| 79 | EGF | 1418093_a_at | 10553 | 0.072 | -0.1891 | No | ||
| 80 | EDA2R | 1440085_at | 10859 | 0.063 | -0.2026 | No | ||
| 81 | TGFA | 1421942_s_at 1450421_at | 10896 | 0.062 | -0.2038 | No | ||
| 82 | RIPK2 | 1421236_at 1450173_at | 11268 | 0.052 | -0.2204 | No | ||
| 83 | ACVR2B | 1419140_at 1439856_at | 11514 | 0.045 | -0.2313 | No | ||
| 84 | EDG2 | 1417143_at 1426110_a_at 1448606_at | 11551 | 0.044 | -0.2326 | No | ||
| 85 | ALS2 | 1417783_at 1417784_at 1431608_at 1446183_at | 11636 | 0.041 | -0.2361 | No | ||
| 86 | FGD4 | 1425037_at 1426041_a_at 1426042_at 1451659_at 1455337_at | 11686 | 0.040 | -0.2380 | No | ||
| 87 | CDH13 | 1423551_at 1431824_at 1454015_a_at 1459535_at | 11817 | 0.036 | -0.2437 | No | ||
| 88 | PLCE1 | 1452398_at | 11966 | 0.031 | -0.2502 | No | ||
| 89 | CANT1 | 1421476_a_at 1433709_at 1451688_s_at | 12008 | 0.030 | -0.2518 | No | ||
| 90 | MYD88 | 1419272_at | 12527 | 0.014 | -0.2755 | No | ||
| 91 | GIT1 | 1454759_at | 12588 | 0.013 | -0.2782 | No | ||
| 92 | FGD2 | 1419515_at | 13335 | -0.012 | -0.3124 | No | ||
| 93 | ZIC1 | 1423477_at 1439627_at | 13344 | -0.012 | -0.3127 | No | ||
| 94 | OTUD7B | 1428953_at 1429139_at 1435552_at 1457716_at | 13548 | -0.019 | -0.3218 | No | ||
| 95 | CDKN2B | 1449152_at | 13553 | -0.019 | -0.3219 | No | ||
| 96 | OTX2 | 1425926_a_at | 13997 | -0.036 | -0.3419 | No | ||
| 97 | FASLG | 1418803_a_at 1449235_at | 14461 | -0.054 | -0.3628 | No | ||
| 98 | BCL10 | 1418970_a_at 1418971_x_at 1418972_at 1443524_x_at | 14546 | -0.057 | -0.3662 | No | ||
| 99 | CDC42BPB | 1434652_at 1459167_at | 14697 | -0.063 | -0.3725 | No | ||
| 100 | MDFI | 1420713_a_at 1425924_at | 14882 | -0.070 | -0.3804 | No | ||
| 101 | FGD6 | 1419322_at 1435467_at | 15034 | -0.077 | -0.3867 | No | ||
| 102 | TNFAIP3 | 1433699_at 1450829_at | 15052 | -0.078 | -0.3868 | No | ||
| 103 | ZDHHC13 | 1428307_at 1460048_at | 15079 | -0.079 | -0.3873 | No | ||
| 104 | HTR2B | 1422125_at | 15171 | -0.085 | -0.3908 | No | ||
| 105 | RGS20 | 1421493_a_at 1443694_at 1446199_at | 15395 | -0.096 | -0.4002 | No | ||
| 106 | RGS11 | 1425245_a_at | 15778 | -0.114 | -0.4168 | No | ||
| 107 | SNX6 | 1425148_a_at 1429497_s_at 1447234_s_at 1451602_at | 15814 | -0.117 | -0.4174 | No | ||
| 108 | SLA2 | 1437504_at 1451492_at | 15815 | -0.117 | -0.4164 | No | ||
| 109 | MIB2 | 1424862_s_at | 15840 | -0.118 | -0.4165 | No | ||
| 110 | HGS | 1416540_at | 15845 | -0.118 | -0.4157 | No | ||
| 111 | REL | 1420710_at | 16007 | -0.128 | -0.4220 | No | ||
| 112 | DMPK | 1422989_a_at 1434944_at | 16010 | -0.129 | -0.4210 | No | ||
| 113 | TRADD | 1429117_at 1439910_a_at 1452622_a_at | 16324 | -0.150 | -0.4341 | No | ||
| 114 | RHOA | 1437628_s_at | 16372 | -0.153 | -0.4350 | No | ||
| 115 | RASGRP4 | 1425380_at | 16421 | -0.157 | -0.4358 | No | ||
| 116 | RIPK1 | 1419508_at 1439273_at 1449485_at | 16494 | -0.162 | -0.4377 | No | ||
| 117 | TRAF6 | 1421376_at 1421377_at 1435350_at 1443288_at 1446940_at 1446977_at | 16565 | -0.168 | -0.4395 | No | ||
| 118 | MAPK8IP1 | 1425679_a_at 1440619_at | 16574 | -0.169 | -0.4385 | No | ||
| 119 | CXXC5 | 1431469_a_at 1448960_at | 16705 | -0.180 | -0.4429 | No | ||
| 120 | RGS16 | 1426037_a_at 1451452_a_at 1455265_a_at | 16761 | -0.184 | -0.4439 | No | ||
| 121 | HIPK2 | 1424863_a_at 1424864_at 1425983_x_at 1428433_at 1429566_a_at 1432523_at 1446167_at 1448631_a_at 1456022_at 1458520_at | 17004 | -0.207 | -0.4532 | No | ||
| 122 | CD40 | 1439221_s_at 1449473_s_at 1460415_a_at | 17136 | -0.221 | -0.4574 | No | ||
| 123 | SLC44A2 | 1428065_at 1438559_x_at 1438860_a_at | 17396 | -0.249 | -0.4672 | No | ||
| 124 | INHBA | 1422053_at 1458291_at | 17416 | -0.252 | -0.4659 | No | ||
| 125 | FGD1 | 1416865_at 1437369_at 1459278_at | 17723 | -0.287 | -0.4775 | No | ||
| 126 | IL31RA | 1445095_at 1451535_at | 17911 | -0.312 | -0.4834 | No | ||
| 127 | TRAF7 | 1424320_a_at | 18034 | -0.328 | -0.4862 | No | ||
| 128 | SNF1LK2 | 1444472_at 1457276_at | 18097 | -0.338 | -0.4862 | No | ||
| 129 | GNG4 | 1417943_at 1417944_at 1447669_s_at | 18181 | -0.350 | -0.4871 | No | ||
| 130 | SIGIRR | 1449163_at | 18196 | -0.352 | -0.4847 | No | ||
| 131 | MALT1 | 1445068_at 1456126_at 1456429_at | 18297 | -0.366 | -0.4862 | No | ||
| 132 | THY1 | 1423135_at | 18309 | -0.368 | -0.4835 | No | ||
| 133 | TBK1 | 1422469_at 1445571_at 1460315_s_at | 18339 | -0.371 | -0.4817 | No | ||
| 134 | RALBP1 | 1417248_at 1448634_at | 18424 | -0.383 | -0.4823 | No | ||
| 135 | BRAP | 1416738_at 1416739_a_at | 18747 | -0.444 | -0.4933 | No | ||
| 136 | ENG | 1417271_a_at 1432176_a_at | 18758 | -0.447 | -0.4900 | No | ||
| 137 | CLCF1 | 1437270_a_at 1437271_at 1450262_at | 18800 | -0.456 | -0.4880 | No | ||
| 138 | MAP3K7IP2 | 1423462_at 1451003_at | 18957 | -0.490 | -0.4910 | No | ||
| 139 | TICAM1 | 1454676_s_at | 19120 | -0.525 | -0.4939 | Yes | ||
| 140 | SMURF1 | 1443393_at | 19172 | -0.538 | -0.4917 | Yes | ||
| 141 | TMEM9B | 1417874_at 1448867_at | 19177 | -0.540 | -0.4873 | Yes | ||
| 142 | MDFIC | 1427040_at | 19282 | -0.568 | -0.4872 | Yes | ||
| 143 | TSC1 | 1422043_at 1439989_at 1455252_at | 19367 | -0.587 | -0.4860 | Yes | ||
| 144 | TMEM101 | 1451315_at | 19475 | -0.613 | -0.4857 | Yes | ||
| 145 | RGS4 | 1416286_at 1416287_at 1448285_at | 19529 | -0.626 | -0.4828 | Yes | ||
| 146 | SCOTIN | 1423986_a_at 1437503_a_at | 19580 | -0.640 | -0.4797 | Yes | ||
| 147 | NDFIP2 | 1452066_a_at | 19587 | -0.641 | -0.4745 | Yes | ||
| 148 | ARHGAP27 | 1416758_at 1457655_x_at | 19637 | -0.658 | -0.4711 | Yes | ||
| 149 | RELA | 1419536_a_at | 19723 | -0.683 | -0.4692 | Yes | ||
| 150 | SLC35B2 | 1423927_at 1429007_at | 19725 | -0.684 | -0.4634 | Yes | ||
| 151 | CENTD2 | 1428064_at | 19735 | -0.687 | -0.4580 | Yes | ||
| 152 | CDC42BPG | 1436197_at | 19840 | -0.721 | -0.4566 | Yes | ||
| 153 | RAMP1 | 1417481_at 1447474_at | 19846 | -0.724 | -0.4506 | Yes | ||
| 154 | F2R | 1437308_s_at 1450852_s_at | 19855 | -0.727 | -0.4448 | Yes | ||
| 155 | HCLS1 | 1418842_at | 19868 | -0.733 | -0.4391 | Yes | ||
| 156 | SLC20A1 | 1438824_at 1446441_at 1448568_a_at | 19943 | -0.759 | -0.4360 | Yes | ||
| 157 | FADD | 1416888_at 1446499_at | 20011 | -0.786 | -0.4324 | Yes | ||
| 158 | BIRC2 | 1418854_at | 20095 | -0.816 | -0.4292 | Yes | ||
| 159 | PTEN | 1422553_at 1441593_at 1444325_at 1450655_at 1454722_at 1455728_at 1457493_at | 20113 | -0.822 | -0.4230 | Yes | ||
| 160 | GRK6 | 1437436_s_at 1450480_a_at 1451672_at | 20236 | -0.862 | -0.4213 | Yes | ||
| 161 | GPR177 | 1423824_at 1423825_at 1437434_a_at 1458319_at | 20312 | -0.897 | -0.4171 | Yes | ||
| 162 | MAP3K3 | 1426686_s_at 1426687_at 1436522_at | 20318 | -0.899 | -0.4096 | Yes | ||
| 163 | RGS3 | 1425701_a_at 1427648_at 1449516_a_at 1454026_a_at | 20320 | -0.900 | -0.4020 | Yes | ||
| 164 | NOTCH2 | 1451889_at 1455556_at | 20365 | -0.915 | -0.3962 | Yes | ||
| 165 | NUP62 | 1415925_a_at 1415926_at 1437612_at 1438917_x_at 1447905_x_at 1448150_at | 20398 | -0.933 | -0.3897 | Yes | ||
| 166 | ADRB2 | 1437302_at | 20462 | -0.963 | -0.3844 | Yes | ||
| 167 | ACVR1 | 1416786_at 1416787_at 1448460_at 1457551_at | 20596 | -1.046 | -0.3816 | Yes | ||
| 168 | FLNA | 1426677_at | 20659 | -1.088 | -0.3752 | Yes | ||
| 169 | TRAT1 | 1427532_at 1437561_at | 20681 | -1.103 | -0.3667 | Yes | ||
| 170 | ARF6 | 1418822_a_at 1418823_at 1418824_at | 20682 | -1.104 | -0.3573 | Yes | ||
| 171 | TAX1BP3 | 1424169_at | 20688 | -1.108 | -0.3481 | Yes | ||
| 172 | ACVR1B | 1422098_at 1433725_at | 20742 | -1.142 | -0.3408 | Yes | ||
| 173 | RGS2 | 1419247_at 1419248_at 1447830_s_at | 20858 | -1.223 | -0.3356 | Yes | ||
| 174 | IKBKE | 1417813_at | 20898 | -1.262 | -0.3266 | Yes | ||
| 175 | FGD3 | 1433398_at 1450235_at 1454593_at | 20975 | -1.326 | -0.3188 | Yes | ||
| 176 | CC2D1A | 1426422_at | 20990 | -1.342 | -0.3080 | Yes | ||
| 177 | TAOK3 | 1435964_a_at 1444366_at 1444881_at 1447257_at 1455733_at | 21028 | -1.394 | -0.2978 | Yes | ||
| 178 | RHOH | 1429319_at 1441385_at 1443015_at | 21077 | -1.451 | -0.2876 | Yes | ||
| 179 | SOCS1 | 1440047_at 1450446_a_at | 21129 | -1.501 | -0.2772 | Yes | ||
| 180 | SQSTM1 | 1440076_at 1442237_at 1444021_at 1450957_a_at | 21130 | -1.503 | -0.2643 | Yes | ||
| 181 | CENTD3 | 1419833_s_at 1447407_at 1451282_at | 21133 | -1.505 | -0.2516 | Yes | ||
| 182 | RGS12 | 1429380_at 1453129_a_at 1457511_at 1459566_at | 21134 | -1.506 | -0.2387 | Yes | ||
| 183 | SMAD4 | 1422485_at 1422486_a_at 1422487_at 1444205_at | 21192 | -1.576 | -0.2279 | Yes | ||
| 184 | RGS14 | 1419221_a_at | 21230 | -1.635 | -0.2157 | Yes | ||
| 185 | CFLAR | 1424996_at 1425686_at 1425687_at 1439600_at 1449317_at | 21351 | -1.795 | -0.2059 | Yes | ||
| 186 | TNFRSF1A | 1417291_at | 21427 | -1.934 | -0.1928 | Yes | ||
| 187 | NLK | 1419112_at 1435970_at 1443279_at 1445427_at 1456467_s_at | 21448 | -1.981 | -0.1768 | Yes | ||
| 188 | TAOK2 | 1438208_at 1443642_at 1456826_at | 21488 | -2.062 | -0.1610 | Yes | ||
| 189 | ATP2C1 | 1431196_at 1434386_at 1437738_at 1439215_at 1442742_at | 21557 | -2.215 | -0.1453 | Yes | ||
| 190 | EEF1E1 | 1444757_at 1449044_at | 21736 | -2.879 | -0.1289 | Yes | ||
| 191 | DTX1 | 1425822_a_at 1458643_at | 21771 | -3.048 | -0.1044 | Yes | ||
| 192 | GIT2 | 1423391_at 1432160_at 1435925_at | 21792 | -3.156 | -0.0784 | Yes | ||
| 193 | MIER1 | 1433755_at 1443400_at 1443879_at 1453760_at | 21816 | -3.301 | -0.0513 | Yes | ||
| 194 | FKBP1A | 1416036_at 1438795_x_at 1438958_x_at 1448184_at 1456196_x_at | 21931 | -6.642 | 0.0001 | Yes |

