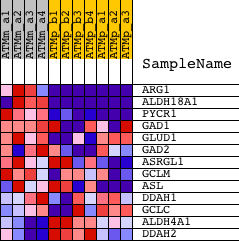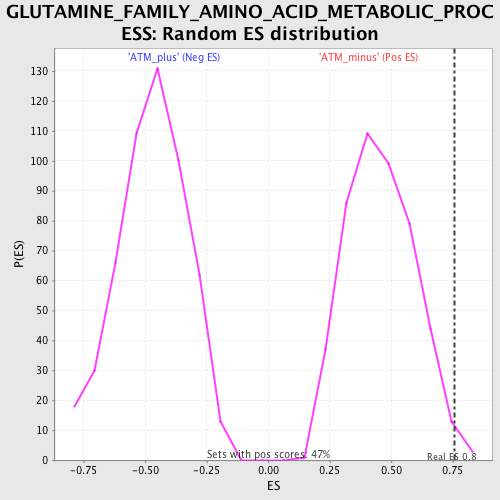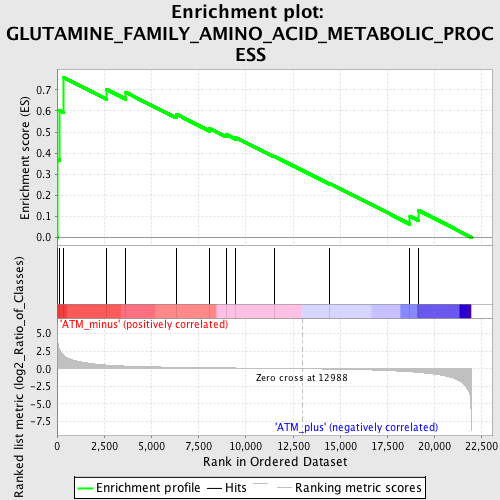
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | GLUTAMINE_FAMILY_AMINO_ACID_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.7585497 |
| Normalized Enrichment Score (NES) | 1.6611878 |
| Nominal p-value | 0.008492569 |
| FDR q-value | 0.20078313 |
| FWER p-Value | 0.961 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARG1 | 1419549_at | 19 | 4.174 | 0.3710 | Yes | ||
| 2 | ALDH18A1 | 1415836_at 1435855_x_at 1437325_x_at 1437333_x_at 1437620_x_at | 136 | 2.652 | 0.6020 | Yes | ||
| 3 | PYCR1 | 1424556_at | 357 | 1.870 | 0.7585 | Yes | ||
| 4 | GAD1 | 1416561_at 1416562_at | 2626 | 0.519 | 0.7013 | No | ||
| 5 | GLUD1 | 1416209_at 1448253_at | 3631 | 0.373 | 0.6887 | No | ||
| 6 | GAD2 | 1421978_at | 6308 | 0.208 | 0.5852 | No | ||
| 7 | ASRGL1 | 1424395_at 1424396_a_at 1460360_at | 8081 | 0.146 | 0.5174 | No | ||
| 8 | GCLM | 1418627_at | 8970 | 0.118 | 0.4874 | No | ||
| 9 | ASL | 1448350_at | 9435 | 0.104 | 0.4755 | No | ||
| 10 | DDAH1 | 1429298_at 1429299_at 1434519_at 1438879_at 1454995_at 1455400_at | 11497 | 0.045 | 0.3855 | No | ||
| 11 | GCLC | 1424296_at | 14439 | -0.052 | 0.2561 | No | ||
| 12 | ALDH4A1 | 1452375_at | 18686 | -0.431 | 0.1008 | No | ||
| 13 | DDAH2 | 1416457_at | 19146 | -0.531 | 0.1272 | No |