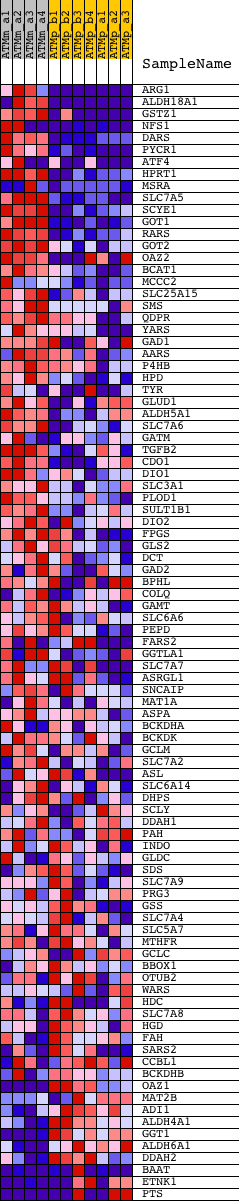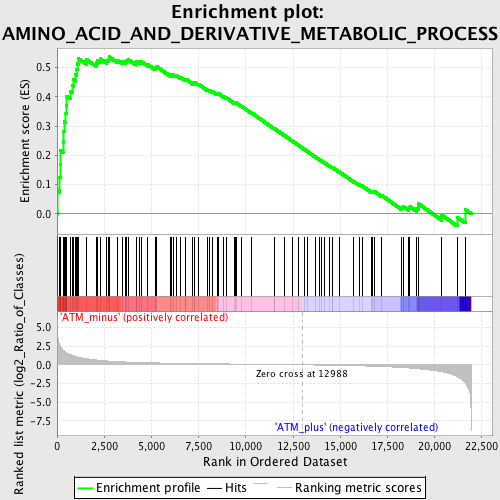
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | AMINO_ACID_AND_DERIVATIVE_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.53668255 |
| Normalized Enrichment Score (NES) | 1.7523875 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.15943824 |
| FWER p-Value | 0.655 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ARG1 | 1419549_at | 19 | 4.174 | 0.0792 | Yes | ||
| 2 | ALDH18A1 | 1415836_at 1435855_x_at 1437325_x_at 1437333_x_at 1437620_x_at | 136 | 2.652 | 0.1247 | Yes | ||
| 3 | GSTZ1 | 1427552_a_at 1458134_at | 186 | 2.447 | 0.1694 | Yes | ||
| 4 | NFS1 | 1416373_at 1431431_a_at 1442017_at | 190 | 2.422 | 0.2157 | Yes | ||
| 5 | DARS | 1423800_at | 319 | 1.933 | 0.2469 | Yes | ||
| 6 | PYCR1 | 1424556_at | 357 | 1.870 | 0.2811 | Yes | ||
| 7 | ATF4 | 1438992_x_at 1439258_at 1448135_at | 378 | 1.823 | 0.3151 | Yes | ||
| 8 | HPRT1 | 1448736_a_at | 464 | 1.663 | 0.3431 | Yes | ||
| 9 | MSRA | 1442385_at 1448856_a_at 1458923_at | 504 | 1.590 | 0.3718 | Yes | ||
| 10 | SLC7A5 | 1418326_at | 515 | 1.580 | 0.4016 | Yes | ||
| 11 | SCYE1 | 1416486_at | 700 | 1.362 | 0.4193 | Yes | ||
| 12 | GOT1 | 1450970_at | 795 | 1.271 | 0.4394 | Yes | ||
| 13 | RARS | 1416312_at | 855 | 1.222 | 0.4601 | Yes | ||
| 14 | GOT2 | 1417715_a_at 1417716_at | 993 | 1.127 | 0.4755 | Yes | ||
| 15 | OAZ2 | 1426763_at 1426764_at 1442541_at 1442735_at | 1045 | 1.091 | 0.4940 | Yes | ||
| 16 | BCAT1 | 1430111_a_at 1450871_a_at | 1077 | 1.071 | 0.5132 | Yes | ||
| 17 | MCCC2 | 1428021_at 1432472_a_at 1454840_at | 1141 | 1.036 | 0.5301 | Yes | ||
| 18 | SLC25A15 | 1420966_at 1420967_at | 1541 | 0.834 | 0.5279 | Yes | ||
| 19 | SMS | 1428699_at 1434190_at | 2072 | 0.647 | 0.5160 | Yes | ||
| 20 | QDPR | 1423664_at 1437993_x_at | 2150 | 0.628 | 0.5245 | Yes | ||
| 21 | YARS | 1460638_at | 2288 | 0.591 | 0.5296 | Yes | ||
| 22 | GAD1 | 1416561_at 1416562_at | 2626 | 0.519 | 0.5241 | Yes | ||
| 23 | AARS | 1423685_at 1451083_s_at | 2724 | 0.499 | 0.5292 | Yes | ||
| 24 | P4HB | 1437465_a_at 1444326_at | 2767 | 0.489 | 0.5367 | Yes | ||
| 25 | HPD | 1424618_at | 3192 | 0.423 | 0.5254 | No | ||
| 26 | TYR | 1417717_a_at 1448821_at 1456095_at | 3473 | 0.390 | 0.5201 | No | ||
| 27 | GLUD1 | 1416209_at 1448253_at | 3631 | 0.373 | 0.5200 | No | ||
| 28 | ALDH5A1 | 1453065_at | 3698 | 0.364 | 0.5240 | No | ||
| 29 | SLC7A6 | 1433467_at 1460541_at | 3790 | 0.355 | 0.5266 | No | ||
| 30 | GATM | 1423569_at | 4180 | 0.322 | 0.5150 | No | ||
| 31 | TGFB2 | 1423250_a_at 1438303_at 1446141_at 1450922_a_at 1450923_at | 4218 | 0.320 | 0.5194 | No | ||
| 32 | CDO1 | 1448842_at | 4344 | 0.311 | 0.5197 | No | ||
| 33 | DIO1 | 1417991_at | 4444 | 0.304 | 0.5210 | No | ||
| 34 | SLC3A1 | 1448741_at | 4810 | 0.280 | 0.5096 | No | ||
| 35 | PLOD1 | 1416289_at 1445893_at | 5188 | 0.259 | 0.4973 | No | ||
| 36 | SULT1B1 | 1418940_at | 5235 | 0.257 | 0.5002 | No | ||
| 37 | DIO2 | 1418937_at 1418938_at 1426081_a_at | 5268 | 0.256 | 0.5036 | No | ||
| 38 | FPGS | 1460673_at | 5994 | 0.221 | 0.4746 | No | ||
| 39 | GLS2 | 1435245_at | 6074 | 0.218 | 0.4752 | No | ||
| 40 | DCT | 1418028_at | 6174 | 0.213 | 0.4748 | No | ||
| 41 | GAD2 | 1421978_at | 6308 | 0.208 | 0.4727 | No | ||
| 42 | BPHL | 1420235_at 1424242_at 1430531_at | 6509 | 0.200 | 0.4673 | No | ||
| 43 | COLQ | 1448081_at | 6776 | 0.190 | 0.4588 | No | ||
| 44 | GAMT | 1422558_at | 6822 | 0.189 | 0.4604 | No | ||
| 45 | SLC6A6 | 1420148_at 1421346_a_at 1437149_at 1449751_at | 7192 | 0.175 | 0.4468 | No | ||
| 46 | PEPD | 1416712_at | 7266 | 0.173 | 0.4468 | No | ||
| 47 | FARS2 | 1431354_a_at 1439406_x_at | 7295 | 0.172 | 0.4488 | No | ||
| 48 | GGTLA1 | 1418216_at 1439420_x_at 1455747_at | 7476 | 0.166 | 0.4438 | No | ||
| 49 | SLC7A7 | 1417392_a_at 1447181_s_at | 7984 | 0.149 | 0.4234 | No | ||
| 50 | ASRGL1 | 1424395_at 1424396_a_at 1460360_at | 8081 | 0.146 | 0.4218 | No | ||
| 51 | SNCAIP | 1423499_at 1430463_a_at 1441037_at | 8218 | 0.142 | 0.4183 | No | ||
| 52 | MAT1A | 1423147_at | 8477 | 0.134 | 0.4091 | No | ||
| 53 | ASPA | 1418472_at | 8515 | 0.133 | 0.4099 | No | ||
| 54 | BCKDHA | 1416647_at | 8542 | 0.132 | 0.4113 | No | ||
| 55 | BCKDK | 1443813_x_at 1460644_at | 8831 | 0.122 | 0.4004 | No | ||
| 56 | GCLM | 1418627_at | 8970 | 0.118 | 0.3964 | No | ||
| 57 | SLC7A2 | 1422648_at 1426008_a_at 1436555_at 1440506_at 1450703_at | 9381 | 0.106 | 0.3797 | No | ||
| 58 | ASL | 1448350_at | 9435 | 0.104 | 0.3792 | No | ||
| 59 | SLC6A14 | 1420503_at 1420504_at | 9476 | 0.103 | 0.3794 | No | ||
| 60 | DHPS | 1434003_a_at 1434004_at | 9773 | 0.095 | 0.3676 | No | ||
| 61 | SCLY | 1417671_at 1441192_at | 10312 | 0.079 | 0.3445 | No | ||
| 62 | DDAH1 | 1429298_at 1429299_at 1434519_at 1438879_at 1454995_at 1455400_at | 11497 | 0.045 | 0.2912 | No | ||
| 63 | PAH | 1454638_a_at | 11537 | 0.044 | 0.2903 | No | ||
| 64 | INDO | 1420437_at | 12066 | 0.028 | 0.2666 | No | ||
| 65 | GLDC | 1416049_at 1459959_at | 12463 | 0.016 | 0.2488 | No | ||
| 66 | SDS | 1424744_at | 12776 | 0.007 | 0.2347 | No | ||
| 67 | SLC7A9 | 1448783_at | 13106 | -0.004 | 0.2197 | No | ||
| 68 | PRG3 | 1449924_at | 13266 | -0.010 | 0.2126 | No | ||
| 69 | GSS | 1441931_x_at 1448273_at | 13704 | -0.025 | 0.1931 | No | ||
| 70 | SLC7A4 | 1426068_at 1426069_s_at 1436776_x_at | 13919 | -0.033 | 0.1839 | No | ||
| 71 | SLC5A7 | 1421428_at | 14024 | -0.037 | 0.1798 | No | ||
| 72 | MTHFR | 1422132_at 1434087_at 1450498_at | 14143 | -0.042 | 0.1752 | No | ||
| 73 | GCLC | 1424296_at | 14439 | -0.052 | 0.1627 | No | ||
| 74 | BBOX1 | 1419618_at | 14564 | -0.058 | 0.1581 | No | ||
| 75 | OTUB2 | 1417575_at 1417576_a_at | 14589 | -0.058 | 0.1582 | No | ||
| 76 | WARS | 1415694_at 1425106_a_at 1434813_x_at 1437832_x_at | 14931 | -0.072 | 0.1439 | No | ||
| 77 | HDC | 1451796_s_at 1454713_s_at | 15711 | -0.111 | 0.1104 | No | ||
| 78 | SLC7A8 | 1417929_at | 15987 | -0.127 | 0.1002 | No | ||
| 79 | HGD | 1452986_at | 16155 | -0.138 | 0.0952 | No | ||
| 80 | FAH | 1417220_at | 16635 | -0.174 | 0.0767 | No | ||
| 81 | SARS2 | 1448522_at | 16708 | -0.180 | 0.0768 | No | ||
| 82 | CCBL1 | 1428151_x_at 1438348_x_at 1446302_at 1452678_a_at | 16834 | -0.191 | 0.0747 | No | ||
| 83 | BCKDHB | 1427153_at | 17183 | -0.226 | 0.0631 | No | ||
| 84 | OAZ1 | 1428868_a_at 1436292_a_at | 18219 | -0.355 | 0.0225 | No | ||
| 85 | MAT2B | 1448196_at | 18318 | -0.369 | 0.0251 | No | ||
| 86 | ADI1 | 1449076_x_at | 18634 | -0.420 | 0.0188 | No | ||
| 87 | ALDH4A1 | 1452375_at | 18686 | -0.431 | 0.0247 | No | ||
| 88 | GGT1 | 1448485_at | 19029 | -0.505 | 0.0187 | No | ||
| 89 | ALDH6A1 | 1448104_at | 19132 | -0.529 | 0.0242 | No | ||
| 90 | DDAH2 | 1416457_at | 19146 | -0.531 | 0.0338 | No | ||
| 91 | BAAT | 1419001_at 1419002_s_at | 20376 | -0.921 | -0.0048 | No | ||
| 92 | ETNK1 | 1419884_at 1430996_at 1433514_at 1433515_s_at 1439972_at 1449650_at 1454633_at | 21184 | -1.566 | -0.0117 | No | ||
| 93 | PTS | 1450660_at 1458402_at | 21618 | -2.400 | 0.0145 | No |