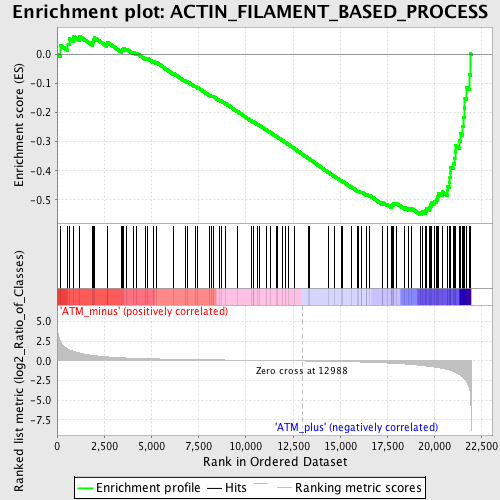
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | ACTIN_FILAMENT_BASED_PROCESS |
| Enrichment Score (ES) | -0.549962 |
| Normalized Enrichment Score (NES) | -1.6801206 |
| Nominal p-value | 0.0015360983 |
| FDR q-value | 0.29574877 |
| FWER p-Value | 0.939 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ABL1 | 1423999_at 1441291_at 1444134_at 1445153_at | 163 | 2.553 | 0.0310 | No | ||
| 2 | SSH1 | 1433874_at 1438253_at 1439237_a_at 1455854_a_at | 575 | 1.510 | 0.0350 | No | ||
| 3 | MYH9 | 1417472_at 1420170_at 1420171_s_at 1420172_at 1440708_at | 640 | 1.418 | 0.0534 | No | ||
| 4 | DSTN | 1417124_at | 883 | 1.194 | 0.0603 | No | ||
| 5 | ARHGEF10L | 1460405_at | 1179 | 1.013 | 0.0621 | No | ||
| 6 | RHOJ | 1418892_at 1444982_at | 1884 | 0.706 | 0.0405 | No | ||
| 7 | DLG1 | 1415691_at 1445798_at 1450768_at 1459635_at | 1930 | 0.690 | 0.0488 | No | ||
| 8 | GSN | 1415812_at 1436991_x_at 1437171_x_at 1441225_at 1456312_x_at 1456568_at 1456569_x_at | 1975 | 0.677 | 0.0570 | No | ||
| 9 | RASA1 | 1426476_at 1426477_at 1426478_at 1438998_at | 2641 | 0.515 | 0.0343 | No | ||
| 10 | DBN1 | 1426024_a_at 1451734_a_at | 2688 | 0.505 | 0.0398 | No | ||
| 11 | ADRA2A | 1423022_at 1433600_at 1433601_at | 3400 | 0.399 | 0.0133 | No | ||
| 12 | ELMO1 | 1424523_at 1446610_at 1450208_a_at | 3445 | 0.393 | 0.0172 | No | ||
| 13 | FSCN2 | 1440605_at | 3535 | 0.384 | 0.0189 | No | ||
| 14 | NCK1 | 1421487_a_at 1424543_at 1447271_at | 3679 | 0.366 | 0.0179 | No | ||
| 15 | MYO9B | 1418031_at 1438533_at 1459110_at | 4021 | 0.335 | 0.0073 | No | ||
| 16 | VIL1 | 1448837_at | 4208 | 0.320 | 0.0036 | No | ||
| 17 | LATS1 | 1427679_at 1460264_at | 4699 | 0.287 | -0.0145 | No | ||
| 18 | CRK | 1416201_at 1425855_a_at 1436835_at 1448248_at 1460176_at | 4804 | 0.281 | -0.0151 | No | ||
| 19 | MRAS | 1449590_a_at | 5081 | 0.265 | -0.0237 | No | ||
| 20 | NF1 | 1438067_at 1441523_at 1443097_at 1452525_a_at | 5271 | 0.256 | -0.0285 | No | ||
| 21 | NCK2 | 1416796_at 1416797_at 1458432_at | 6147 | 0.214 | -0.0654 | No | ||
| 22 | RAC1 | 1423734_at 1437674_at 1451086_s_at | 6772 | 0.190 | -0.0911 | No | ||
| 23 | DYNLL1 | 1448682_at | 6892 | 0.186 | -0.0937 | No | ||
| 24 | CDC42BPA | 1435847_at 1436380_at 1438449_at 1442260_at 1446945_at | 7303 | 0.172 | -0.1099 | No | ||
| 25 | ARPC1A | 1416079_a_at | 7445 | 0.167 | -0.1138 | No | ||
| 26 | CXCL12 | 1417574_at 1439084_at 1448823_at | 8090 | 0.145 | -0.1411 | No | ||
| 27 | TNXB | 1450798_at | 8198 | 0.142 | -0.1439 | No | ||
| 28 | MYO6 | 1421120_at 1433942_at 1435559_at | 8260 | 0.141 | -0.1446 | No | ||
| 29 | NF2 | 1421820_a_at 1427708_a_at 1450382_at 1451829_a_at | 8584 | 0.131 | -0.1574 | No | ||
| 30 | SORBS3 | 1419329_at | 8690 | 0.127 | -0.1603 | No | ||
| 31 | ARHGEF2 | 1421042_at 1421043_s_at 1427646_a_at | 8925 | 0.120 | -0.1692 | No | ||
| 32 | LIMA1 | 1422499_at 1425654_a_at 1435726_at 1435727_s_at 1450629_at | 9537 | 0.101 | -0.1957 | No | ||
| 33 | LIMK1 | 1417627_a_at 1425836_a_at 1456234_at 1458989_at | 10271 | 0.080 | -0.2280 | No | ||
| 34 | RND3 | 1416700_at 1416701_at | 10379 | 0.077 | -0.2318 | No | ||
| 35 | FGD5 | 1460578_at | 10393 | 0.077 | -0.2312 | No | ||
| 36 | RAC3 | 1420554_a_at | 10613 | 0.070 | -0.2402 | No | ||
| 37 | GHSR | 1446756_at | 10735 | 0.067 | -0.2447 | No | ||
| 38 | PLEK2 | 1449424_at | 11067 | 0.058 | -0.2590 | No | ||
| 39 | CFL1 | 1448346_at 1455138_x_at | 11312 | 0.051 | -0.2694 | No | ||
| 40 | WASF3 | 1425061_at 1444243_at | 11631 | 0.041 | -0.2834 | No | ||
| 41 | FGD4 | 1425037_at 1426041_a_at 1426042_at 1451659_at 1455337_at | 11686 | 0.040 | -0.2852 | No | ||
| 42 | PRKG1 | 1443663_at 1443811_at 1443812_x_at 1444232_at 1445807_at 1445998_at 1449876_at | 11939 | 0.032 | -0.2963 | No | ||
| 43 | CDC42EP5 | 1418712_at 1430018_at | 12113 | 0.027 | -0.3038 | No | ||
| 44 | GHRL | 1448980_at | 12273 | 0.022 | -0.3108 | No | ||
| 45 | MYH11 | 1418122_at 1448962_at | 12590 | 0.012 | -0.3251 | No | ||
| 46 | FGD2 | 1419515_at | 13335 | -0.012 | -0.3590 | No | ||
| 47 | DST | 1421117_at 1421276_a_at 1423626_at 1442395_at 1446943_at 1450119_at 1458075_at 1459098_at 1459356_at | 13356 | -0.013 | -0.3597 | No | ||
| 48 | TTN | 1427445_a_at 1427446_s_at 1431928_at 1444083_at 1444638_at 1446450_at | 14389 | -0.050 | -0.4062 | No | ||
| 49 | CDC42BPB | 1434652_at 1459167_at | 14697 | -0.063 | -0.4193 | No | ||
| 50 | FGD6 | 1419322_at 1435467_at | 15034 | -0.077 | -0.4335 | No | ||
| 51 | TNNT2 | 1418726_a_at 1424967_x_at 1440424_at | 15127 | -0.082 | -0.4365 | No | ||
| 52 | CXCL1 | 1419209_at 1441855_x_at 1457644_s_at | 15565 | -0.103 | -0.4550 | No | ||
| 53 | BCAR1 | 1439388_s_at 1450622_at | 15932 | -0.124 | -0.4699 | No | ||
| 54 | RND1 | 1455197_at | 15964 | -0.126 | -0.4694 | No | ||
| 55 | ROCK1 | 1423444_at 1423445_at 1441162_at 1446518_at 1450994_at 1460729_at | 16107 | -0.135 | -0.4739 | No | ||
| 56 | NUAK2 | 1429049_at | 16144 | -0.137 | -0.4734 | No | ||
| 57 | RHOA | 1437628_s_at | 16372 | -0.153 | -0.4815 | No | ||
| 58 | ARPC4 | 1423588_at 1423589_at | 16393 | -0.155 | -0.4801 | No | ||
| 59 | WASL | 1426776_at 1426777_a_at 1432155_at 1439832_at 1452193_a_at | 16548 | -0.167 | -0.4847 | No | ||
| 60 | WASF1 | 1418545_at 1440438_at | 17217 | -0.229 | -0.5118 | No | ||
| 61 | PDPK1 | 1415729_at 1416501_at 1459775_at 1459776_x_at | 17246 | -0.231 | -0.5096 | No | ||
| 62 | MYL6 | 1435041_at | 17474 | -0.257 | -0.5161 | No | ||
| 63 | ARPC5 | 1444568_at 1448129_at | 17712 | -0.286 | -0.5227 | No | ||
| 64 | FGD1 | 1416865_at 1437369_at 1459278_at | 17723 | -0.287 | -0.5188 | No | ||
| 65 | MYOZ1 | 1448636_at 1460202_at | 17763 | -0.292 | -0.5162 | No | ||
| 66 | MYO1E | 1420159_at 1420160_s_at 1428509_at 1449753_at 1449941_at | 17800 | -0.296 | -0.5133 | No | ||
| 67 | MYL6B | 1430661_at | 17840 | -0.301 | -0.5106 | No | ||
| 68 | CAPG | 1447803_x_at 1450355_a_at | 17979 | -0.322 | -0.5121 | No | ||
| 69 | ARHGEF17 | 1419938_s_at 1419939_at 1433682_at 1442357_at 1447441_at | 18412 | -0.380 | -0.5261 | No | ||
| 70 | SCIN | 1450276_a_at | 18617 | -0.417 | -0.5292 | No | ||
| 71 | SORBS1 | 1417358_s_at 1425826_a_at 1428471_at 1436737_a_at 1440311_at 1442401_at 1443983_at 1455967_at | 18756 | -0.446 | -0.5288 | No | ||
| 72 | FSCN1 | 1416514_a_at 1416515_at 1416516_at 1444398_at 1448378_at 1457739_at | 19220 | -0.550 | -0.5417 | Yes | ||
| 73 | TSC1 | 1422043_at 1439989_at 1455252_at | 19367 | -0.587 | -0.5395 | Yes | ||
| 74 | ACTA1 | 1427735_a_at | 19508 | -0.620 | -0.5366 | Yes | ||
| 75 | TESK2 | 1426546_at | 19561 | -0.635 | -0.5294 | Yes | ||
| 76 | CENTD2 | 1428064_at | 19735 | -0.687 | -0.5269 | Yes | ||
| 77 | PSCD2 | 1450103_a_at | 19753 | -0.693 | -0.5173 | Yes | ||
| 78 | CDC42BPG | 1436197_at | 19840 | -0.721 | -0.5103 | Yes | ||
| 79 | MYH10 | 1441057_at 1452740_at | 19960 | -0.765 | -0.5043 | Yes | ||
| 80 | SSH2 | 1456153_at | 20098 | -0.818 | -0.4982 | Yes | ||
| 81 | LLGL1 | 1416621_at | 20149 | -0.834 | -0.4879 | Yes | ||
| 82 | CDC42EP2 | 1428750_at 1445870_at | 20185 | -0.844 | -0.4768 | Yes | ||
| 83 | ARHGDIB | 1426454_at | 20392 | -0.931 | -0.4722 | Yes | ||
| 84 | FLNA | 1426677_at | 20659 | -1.088 | -0.4680 | Yes | ||
| 85 | ARF6 | 1418822_a_at 1418823_at 1418824_at | 20682 | -1.104 | -0.4524 | Yes | ||
| 86 | DOCK2 | 1422808_s_at 1437282_at 1438334_at 1459382_at | 20784 | -1.172 | -0.4394 | Yes | ||
| 87 | MYH6 | 1417729_at 1448826_at 1458850_at | 20803 | -1.187 | -0.4223 | Yes | ||
| 88 | KPTN | 1423438_at 1440025_at | 20826 | -1.202 | -0.4052 | Yes | ||
| 89 | WASF2 | 1438683_at | 20834 | -1.206 | -0.3873 | Yes | ||
| 90 | FGD3 | 1433398_at 1450235_at 1454593_at | 20975 | -1.326 | -0.3737 | Yes | ||
| 91 | ARFIP2 | 1424240_at 1435498_at | 21063 | -1.431 | -0.3562 | Yes | ||
| 92 | PLA2G1B | 1416626_at 1433948_at 1433949_x_at 1437015_x_at | 21073 | -1.448 | -0.3347 | Yes | ||
| 93 | NEBL | 1438452_at 1439897_at 1451846_at | 21101 | -1.471 | -0.3138 | Yes | ||
| 94 | RHOF | 1434794_at 1441510_at 1455902_x_at | 21298 | -1.715 | -0.2969 | Yes | ||
| 95 | AMOT | 1425907_s_at 1427584_at 1454890_at | 21372 | -1.832 | -0.2727 | Yes | ||
| 96 | TAOK2 | 1438208_at 1443642_at 1456826_at | 21488 | -2.062 | -0.2469 | Yes | ||
| 97 | FLNB | 1424828_a_at 1426750_at 1442107_at 1445534_at 1458226_at | 21530 | -2.159 | -0.2162 | Yes | ||
| 98 | ATP2C1 | 1431196_at 1434386_at 1437738_at 1439215_at 1442742_at | 21557 | -2.215 | -0.1840 | Yes | ||
| 99 | MTSS1 | 1424826_s_at 1434036_at 1440847_at 1443729_at 1446284_at 1451496_at | 21597 | -2.321 | -0.1508 | Yes | ||
| 100 | CDC42 | 1415724_a_at 1435807_at 1449574_a_at 1460708_s_at | 21696 | -2.726 | -0.1142 | Yes | ||
| 101 | EVL | 1434920_a_at 1440885_at 1445957_at 1450106_a_at 1456453_at 1458889_at | 21831 | -3.401 | -0.0691 | Yes | ||
| 102 | RACGAP1 | 1421546_a_at 1441889_x_at 1451358_a_at | 21907 | -4.892 | 0.0012 | Yes |

