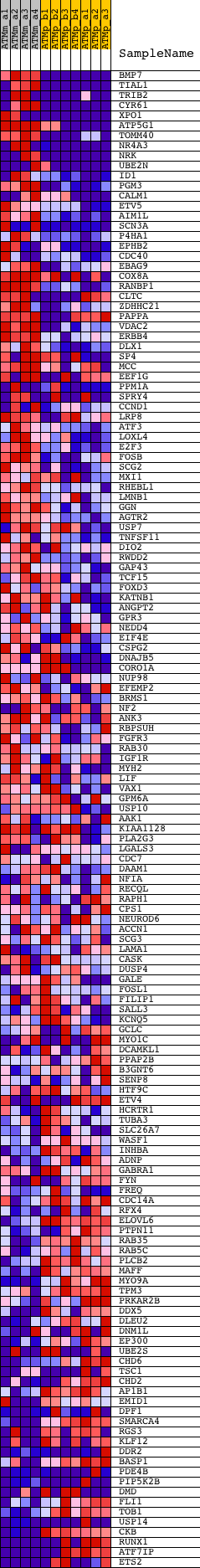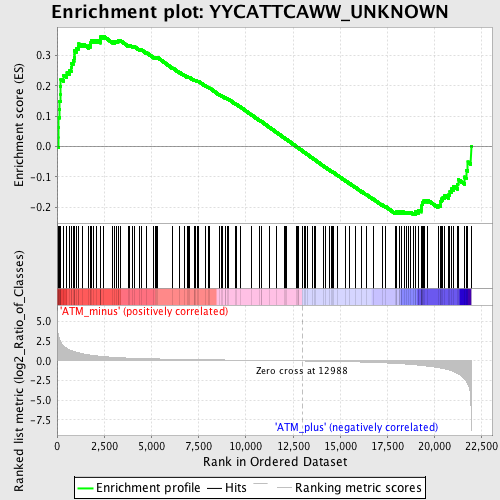
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | YYCATTCAWW_UNKNOWN |
| Enrichment Score (ES) | 0.3637717 |
| Normalized Enrichment Score (NES) | 1.2663798 |
| Nominal p-value | 0.0509915 |
| FDR q-value | 0.28442776 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | BMP7 | 1418910_at 1432410_a_at | 66 | 3.232 | 0.0305 | Yes | ||
| 2 | TIAL1 | 1421148_a_at 1426352_s_at 1446713_at 1452821_at 1455675_a_at 1455676_x_at | 68 | 3.199 | 0.0636 | Yes | ||
| 3 | TRIB2 | 1426640_s_at 1426641_at 1459852_x_at | 77 | 3.168 | 0.0960 | Yes | ||
| 4 | CYR61 | 1416039_x_at 1417848_at 1417849_at 1438133_a_at 1442340_x_at 1457823_at 1459462_at | 126 | 2.725 | 0.1221 | Yes | ||
| 5 | XPO1 | 1418442_at 1418443_at 1448070_at | 141 | 2.630 | 0.1487 | Yes | ||
| 6 | ATP5G1 | 1416020_a_at 1444874_at | 177 | 2.490 | 0.1729 | Yes | ||
| 7 | TOMM40 | 1426118_a_at 1459680_at | 184 | 2.454 | 0.1980 | Yes | ||
| 8 | NR4A3 | 1421079_at 1421080_at | 201 | 2.364 | 0.2218 | Yes | ||
| 9 | NRK | 1436399_s_at 1450078_at 1450079_at | 341 | 1.896 | 0.2350 | Yes | ||
| 10 | UBE2N | 1422559_at 1435384_at | 522 | 1.575 | 0.2431 | Yes | ||
| 11 | ID1 | 1425895_a_at | 674 | 1.389 | 0.2506 | Yes | ||
| 12 | PGM3 | 1428228_at | 745 | 1.311 | 0.2609 | Yes | ||
| 13 | CALM1 | 1417365_a_at 1417366_s_at 1433592_at 1454611_a_at 1455571_x_at | 762 | 1.299 | 0.2737 | Yes | ||
| 14 | ETV5 | 1420998_at 1428142_at 1450082_s_at | 848 | 1.227 | 0.2825 | Yes | ||
| 15 | AIM1L | 1437813_at | 896 | 1.188 | 0.2926 | Yes | ||
| 16 | SCN3A | 1421705_at 1439204_at | 928 | 1.162 | 0.3033 | Yes | ||
| 17 | P4HA1 | 1426519_at 1452094_at | 930 | 1.162 | 0.3152 | Yes | ||
| 18 | EPHB2 | 1425015_at 1425016_at 1442471_at 1445417_at 1454022_at | 1021 | 1.107 | 0.3226 | Yes | ||
| 19 | CDC40 | 1442667_at 1445348_at 1453342_at 1457366_at | 1114 | 1.048 | 0.3292 | Yes | ||
| 20 | EBAG9 | 1422669_at | 1154 | 1.027 | 0.3381 | Yes | ||
| 21 | COX8A | 1416112_at 1436409_at 1448222_x_at | 1367 | 0.911 | 0.3378 | Yes | ||
| 22 | RANBP1 | 1422547_at | 1682 | 0.778 | 0.3314 | Yes | ||
| 23 | CLTC | 1419407_at 1439094_at 1440457_at 1454626_at | 1765 | 0.746 | 0.3354 | Yes | ||
| 24 | ZDHHC21 | 1429488_at | 1788 | 0.736 | 0.3420 | Yes | ||
| 25 | PAPPA | 1427633_a_at 1432591_at 1432592_at 1459375_at | 1802 | 0.732 | 0.3490 | Yes | ||
| 26 | VDAC2 | 1415990_at | 1929 | 0.690 | 0.3504 | Yes | ||
| 27 | ERBB4 | 1427783_at 1446211_at 1447043_at | 2104 | 0.639 | 0.3490 | Yes | ||
| 28 | DLX1 | 1449470_at | 2279 | 0.594 | 0.3472 | Yes | ||
| 29 | SP4 | 1421504_at 1437508_at | 2300 | 0.589 | 0.3524 | Yes | ||
| 30 | MCC | 1438081_at 1442741_at 1442892_at 1459524_at 1459752_at | 2304 | 0.589 | 0.3583 | Yes | ||
| 31 | EEF1G | 1417364_at 1458955_at | 2319 | 0.587 | 0.3638 | Yes | ||
| 32 | PPM1A | 1415678_at 1417221_at 1425537_at 1429500_at 1429501_s_at 1443903_at 1451943_a_at 1453171_s_at | 2476 | 0.550 | 0.3623 | No | ||
| 33 | SPRY4 | 1422020_at 1422021_at 1440867_at 1445669_at | 2944 | 0.459 | 0.3456 | No | ||
| 34 | CCND1 | 1417419_at 1417420_at 1448698_at | 3060 | 0.439 | 0.3449 | No | ||
| 35 | LRP8 | 1421459_a_at 1440882_at 1442347_at | 3141 | 0.429 | 0.3457 | No | ||
| 36 | ATF3 | 1449363_at | 3225 | 0.420 | 0.3462 | No | ||
| 37 | LOXL4 | 1421153_at 1450134_at | 3257 | 0.416 | 0.3491 | No | ||
| 38 | E2F3 | 1427462_at 1434564_at | 3343 | 0.405 | 0.3494 | No | ||
| 39 | FOSB | 1422134_at | 3800 | 0.354 | 0.3321 | No | ||
| 40 | SCG2 | 1450708_at | 3833 | 0.351 | 0.3343 | No | ||
| 41 | MXI1 | 1425732_a_at 1450376_at | 3981 | 0.338 | 0.3311 | No | ||
| 42 | RHEBL1 | 1447818_x_at 1451516_at | 4105 | 0.328 | 0.3288 | No | ||
| 43 | LMNB1 | 1423520_at 1423521_at | 4379 | 0.308 | 0.3195 | No | ||
| 44 | GGN | 1445278_at | 4470 | 0.303 | 0.3185 | No | ||
| 45 | AGTR2 | 1415832_at | 4722 | 0.286 | 0.3099 | No | ||
| 46 | USP7 | 1419920_s_at 1419921_s_at 1434371_x_at 1437118_at 1438249_at 1454948_at 1454949_at | 5124 | 0.263 | 0.2942 | No | ||
| 47 | TNFSF11 | 1419083_at 1451944_a_at | 5201 | 0.259 | 0.2934 | No | ||
| 48 | DIO2 | 1418937_at 1418938_at 1426081_a_at | 5268 | 0.256 | 0.2931 | No | ||
| 49 | RWDD2 | 1428963_at | 5299 | 0.254 | 0.2943 | No | ||
| 50 | GAP43 | 1423537_at 1445567_at | 6123 | 0.216 | 0.2588 | No | ||
| 51 | TCF15 | 1449592_at | 6488 | 0.201 | 0.2441 | No | ||
| 52 | FOXD3 | 1422210_at | 6732 | 0.192 | 0.2350 | No | ||
| 53 | KATNB1 | 1426665_at | 6888 | 0.186 | 0.2298 | No | ||
| 54 | ANGPT2 | 1422415_at 1446903_at 1448831_at | 6968 | 0.183 | 0.2281 | No | ||
| 55 | GPR3 | 1460275_at | 6994 | 0.183 | 0.2288 | No | ||
| 56 | NEDD4 | 1421955_a_at 1444044_at 1450431_a_at 1451109_a_at | 7250 | 0.174 | 0.2189 | No | ||
| 57 | EIF4E | 1450909_at 1457489_at 1459371_at | 7293 | 0.172 | 0.2188 | No | ||
| 58 | CSPG2 | 1421694_a_at 1427256_at 1427257_at 1433043_at 1433044_at 1447586_at 1447887_x_at 1459767_x_at | 7342 | 0.170 | 0.2183 | No | ||
| 59 | DNAJB5 | 1421961_a_at 1421962_at 1450436_s_at | 7441 | 0.167 | 0.2156 | No | ||
| 60 | CORO1A | 1416246_a_at 1435288_at 1455269_a_at | 7481 | 0.166 | 0.2155 | No | ||
| 61 | NUP98 | 1434220_at 1439154_at 1446397_at 1446988_at | 7848 | 0.154 | 0.2003 | No | ||
| 62 | EFEMP2 | 1417018_at 1447668_x_at | 7996 | 0.149 | 0.1951 | No | ||
| 63 | BRMS1 | 1417897_at 1426129_at | 8091 | 0.145 | 0.1923 | No | ||
| 64 | NF2 | 1421820_a_at 1427708_a_at 1450382_at 1451829_a_at | 8584 | 0.131 | 0.1710 | No | ||
| 65 | ANK3 | 1425202_a_at 1439220_at 1447259_at 1451628_a_at 1457288_at | 8695 | 0.127 | 0.1673 | No | ||
| 66 | RBPSUH | 1418114_at 1438848_at 1446436_at 1448957_at 1454896_at | 8779 | 0.124 | 0.1648 | No | ||
| 67 | FGFR3 | 1421841_at 1425796_a_at | 8906 | 0.120 | 0.1603 | No | ||
| 68 | RAB30 | 1426452_a_at | 8919 | 0.120 | 0.1609 | No | ||
| 69 | IGF1R | 1426565_at 1437219_at 1443566_at 1445600_at 1445664_at 1446303_at | 9002 | 0.117 | 0.1584 | No | ||
| 70 | MYH2 | 1425153_at | 9098 | 0.114 | 0.1552 | No | ||
| 71 | LIF | 1421206_at 1421207_at 1450160_at | 9456 | 0.104 | 0.1399 | No | ||
| 72 | VAX1 | 1421774_at | 9504 | 0.102 | 0.1388 | No | ||
| 73 | GPM6A | 1426442_at 1456741_s_at 1459303_at | 9690 | 0.097 | 0.1313 | No | ||
| 74 | USP10 | 1448230_at | 10306 | 0.079 | 0.1039 | No | ||
| 75 | AAK1 | 1420026_at 1435038_s_at 1441782_at | 10729 | 0.067 | 0.0852 | No | ||
| 76 | KIAA1128 | 1431972_a_at 1452223_s_at 1453240_a_at | 10802 | 0.064 | 0.0826 | No | ||
| 77 | PLA2G3 | 1441247_at 1456231_at | 10842 | 0.063 | 0.0815 | No | ||
| 78 | LGALS3 | 1426808_at 1445626_at | 11246 | 0.053 | 0.0635 | No | ||
| 79 | CDC7 | 1426002_a_at 1426021_a_at | 11602 | 0.042 | 0.0477 | No | ||
| 80 | DAAM1 | 1431035_at 1455244_at 1458662_at | 12063 | 0.028 | 0.0268 | No | ||
| 81 | NFIA | 1421162_a_at 1421163_a_at 1427733_a_at 1438236_at 1440660_at 1441547_at 1446742_at 1446990_at 1456087_at 1459431_at | 12110 | 0.027 | 0.0250 | No | ||
| 82 | RECQL | 1418339_at 1425798_a_at 1436371_at 1439047_s_at 1447198_at | 12130 | 0.026 | 0.0244 | No | ||
| 83 | RAPH1 | 1434302_at 1434303_at 1444586_at 1456918_at | 12678 | 0.010 | -0.0006 | No | ||
| 84 | CPS1 | 1455540_at | 12716 | 0.009 | -0.0022 | No | ||
| 85 | NEUROD6 | 1418047_at | 12796 | 0.007 | -0.0058 | No | ||
| 86 | ACCN1 | 1417994_a_at | 12802 | 0.006 | -0.0059 | No | ||
| 87 | SCG3 | 1448628_at | 12975 | 0.000 | -0.0138 | No | ||
| 88 | LAMA1 | 1418153_at | 13115 | -0.004 | -0.0202 | No | ||
| 89 | CASK | 1422518_at 1422519_at 1427692_a_at 1445152_at | 13139 | -0.005 | -0.0212 | No | ||
| 90 | DUSP4 | 1428834_at | 13260 | -0.009 | -0.0266 | No | ||
| 91 | GALE | 1424140_at | 13538 | -0.019 | -0.0391 | No | ||
| 92 | FOSL1 | 1417487_at 1417488_at | 13609 | -0.021 | -0.0421 | No | ||
| 93 | FILIP1 | 1436650_at 1444075_at | 13695 | -0.025 | -0.0457 | No | ||
| 94 | SALL3 | 1441186_at 1452386_at | 14086 | -0.040 | -0.0632 | No | ||
| 95 | KCNQ5 | 1427426_at 1441071_at | 14198 | -0.044 | -0.0679 | No | ||
| 96 | GCLC | 1424296_at | 14439 | -0.052 | -0.0783 | No | ||
| 97 | MYO1C | 1419648_at 1419649_s_at 1449550_at 1449551_at | 14532 | -0.056 | -0.0820 | No | ||
| 98 | DCAMKL1 | 1423125_at 1424270_at 1424271_at 1436659_at 1446190_at 1450863_a_at 1451289_at 1451917_a_at | 14584 | -0.058 | -0.0837 | No | ||
| 99 | PPAP2B | 1429514_at 1446850_at 1448908_at | 14653 | -0.061 | -0.0862 | No | ||
| 100 | B3GNT6 | 1456680_at | 14840 | -0.069 | -0.0940 | No | ||
| 101 | SENP8 | 1437307_at 1438102_at 1454084_a_at | 15283 | -0.090 | -0.1134 | No | ||
| 102 | HTF9C | 1434244_x_at 1441920_x_at 1448114_a_at 1448115_at 1453995_a_at 1457620_at | 15505 | -0.100 | -0.1225 | No | ||
| 103 | ETV4 | 1423232_at 1443381_at | 15791 | -0.115 | -0.1344 | No | ||
| 104 | HCRTR1 | 1436295_at | 16145 | -0.137 | -0.1492 | No | ||
| 105 | TUBA3 | 1438611_at | 16378 | -0.154 | -0.1582 | No | ||
| 106 | SLC26A7 | 1425841_at | 16738 | -0.182 | -0.1728 | No | ||
| 107 | WASF1 | 1418545_at 1440438_at | 17217 | -0.229 | -0.1924 | No | ||
| 108 | INHBA | 1422053_at 1458291_at | 17416 | -0.252 | -0.1988 | No | ||
| 109 | ADNP | 1423069_at | 17917 | -0.314 | -0.2186 | No | ||
| 110 | GABRA1 | 1421280_at 1421281_at 1436889_at 1455766_at | 17954 | -0.319 | -0.2169 | No | ||
| 111 | FYN | 1417558_at 1441647_at 1448765_at | 17958 | -0.320 | -0.2137 | No | ||
| 112 | FREQ | 1434887_at 1450146_at 1460293_at | 18108 | -0.339 | -0.2170 | No | ||
| 113 | CDC14A | 1436913_at 1443184_at 1446493_at 1459517_at | 18135 | -0.342 | -0.2147 | No | ||
| 114 | RFX4 | 1432053_at 1436931_at 1443312_at | 18263 | -0.362 | -0.2168 | No | ||
| 115 | ELOVL6 | 1417403_at 1417404_at 1445062_at 1445578_at 1445641_at | 18264 | -0.362 | -0.2130 | No | ||
| 116 | PTPN11 | 1421196_at 1427699_a_at 1451225_at | 18414 | -0.381 | -0.2159 | No | ||
| 117 | RAB35 | 1433922_at | 18522 | -0.400 | -0.2167 | No | ||
| 118 | RAB5C | 1424684_at | 18632 | -0.420 | -0.2173 | No | ||
| 119 | PLCB2 | 1452481_at | 18698 | -0.433 | -0.2158 | No | ||
| 120 | MAFF | 1418936_at | 18877 | -0.470 | -0.2191 | No | ||
| 121 | MYO9A | 1436307_at 1439361_at 1441567_at 1444541_at 1459991_at | 18974 | -0.494 | -0.2184 | No | ||
| 122 | TPM3 | 1427260_a_at 1427567_a_at 1436958_x_at 1449996_a_at 1449997_at | 18989 | -0.497 | -0.2139 | No | ||
| 123 | PRKAR2B | 1430640_a_at 1438664_at 1456475_s_at | 19135 | -0.529 | -0.2151 | No | ||
| 124 | DDX5 | 1419653_a_at 1423645_a_at 1433809_at 1433810_x_at 1438371_x_at 1454793_x_at | 19138 | -0.529 | -0.2097 | No | ||
| 125 | DLEU2 | 1427410_at 1427411_s_at 1456145_at | 19273 | -0.565 | -0.2100 | No | ||
| 126 | DNM1L | 1428008_at 1428086_at 1428087_at 1452638_s_at | 19293 | -0.571 | -0.2049 | No | ||
| 127 | EP300 | 1434765_at | 19298 | -0.572 | -0.1992 | No | ||
| 128 | UBE2S | 1416726_s_at 1430962_at | 19319 | -0.578 | -0.1941 | No | ||
| 129 | CHD6 | 1427384_at 1431047_at | 19324 | -0.580 | -0.1883 | No | ||
| 130 | TSC1 | 1422043_at 1439989_at 1455252_at | 19367 | -0.587 | -0.1841 | No | ||
| 131 | CHD2 | 1444246_at 1445843_at | 19401 | -0.597 | -0.1794 | No | ||
| 132 | AP1B1 | 1416279_at | 19482 | -0.615 | -0.1767 | No | ||
| 133 | EMID1 | 1442144_at 1449581_at | 19623 | -0.652 | -0.1764 | No | ||
| 134 | DPF1 | 1420529_at 1420530_at 1442440_at | 20174 | -0.839 | -0.1930 | No | ||
| 135 | SMARCA4 | 1426804_at 1426805_at | 20313 | -0.897 | -0.1900 | No | ||
| 136 | RGS3 | 1425701_a_at 1427648_at 1449516_a_at 1454026_a_at | 20320 | -0.900 | -0.1810 | No | ||
| 137 | KLF12 | 1439846_at 1439847_s_at 1441040_at 1455521_at | 20350 | -0.912 | -0.1728 | No | ||
| 138 | DDR2 | 1422738_at 1455688_at | 20433 | -0.949 | -0.1668 | No | ||
| 139 | BASP1 | 1428572_at | 20532 | -1.005 | -0.1609 | No | ||
| 140 | PDE4B | 1422473_at 1422474_at 1442700_at 1444731_at 1445449_at 1446577_at 1447237_at 1447718_at | 20750 | -1.147 | -0.1589 | No | ||
| 141 | PIP5K2B | 1427438_at 1435068_at 1441164_at 1444128_at | 20804 | -1.187 | -0.1491 | No | ||
| 142 | DMD | 1417307_at 1430320_at 1446156_at 1448665_at 1457022_at | 20881 | -1.246 | -0.1396 | No | ||
| 143 | FLI1 | 1422024_at 1433512_at 1441584_at | 21006 | -1.360 | -0.1312 | No | ||
| 144 | TOB1 | 1423176_at 1440844_at | 21205 | -1.593 | -0.1238 | No | ||
| 145 | USP14 | 1416208_at 1437714_x_at 1439201_at 1455829_at | 21231 | -1.636 | -0.1080 | No | ||
| 146 | CKB | 1455106_a_at | 21580 | -2.278 | -0.1004 | No | ||
| 147 | RUNX1 | 1422864_at 1422865_at 1427650_a_at 1427847_at 1440878_at 1444527_at 1446930_at 1452530_a_at 1452531_at 1452578_at 1459470_at | 21665 | -2.559 | -0.0777 | No | ||
| 148 | ATF7IP | 1446323_at 1449192_at 1454973_at 1459096_at | 21759 | -2.998 | -0.0509 | No | ||
| 149 | ETS2 | 1416268_at 1437809_x_at 1447685_x_at | 21919 | -5.684 | 0.0007 | No |