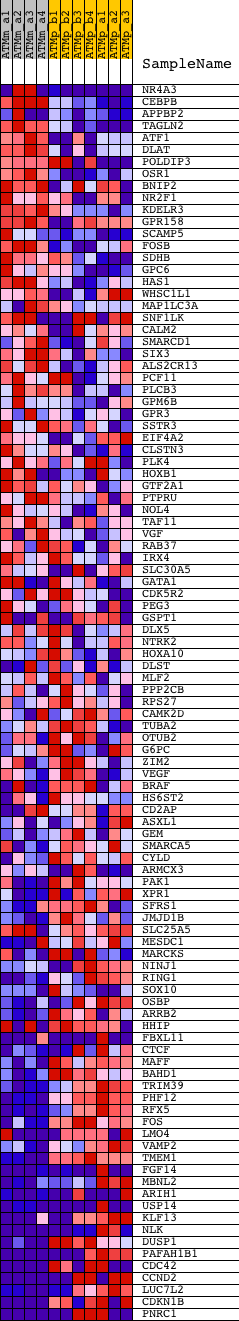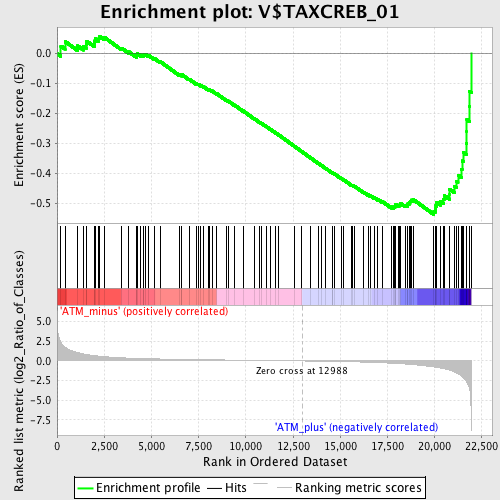
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | V$TAXCREB_01 |
| Enrichment Score (ES) | -0.53718764 |
| Normalized Enrichment Score (NES) | -1.6429783 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.05902165 |
| FWER p-Value | 0.333 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NR4A3 | 1421079_at 1421080_at | 201 | 2.364 | 0.0255 | No | ||
| 2 | CEBPB | 1418901_at 1427844_a_at | 421 | 1.731 | 0.0409 | No | ||
| 3 | APPBP2 | 1430265_at 1434039_at 1434040_at 1434041_at 1444886_at 1447855_x_at 1451251_at | 1087 | 1.066 | 0.0261 | No | ||
| 4 | TAGLN2 | 1426529_a_at | 1392 | 0.895 | 0.0253 | No | ||
| 5 | ATF1 | 1417296_at | 1538 | 0.835 | 0.0309 | No | ||
| 6 | DLAT | 1426264_at 1426265_x_at 1452005_at | 1556 | 0.829 | 0.0423 | No | ||
| 7 | POLDIP3 | 1434176_x_at 1437335_x_at 1437837_x_at 1440022_at 1452709_at 1455788_x_at | 1971 | 0.678 | 0.0333 | No | ||
| 8 | OSR1 | 1449350_at | 1988 | 0.675 | 0.0425 | No | ||
| 9 | BNIP2 | 1422490_at 1422491_a_at 1431731_at 1433025_x_at 1441989_at 1453993_a_at 1454441_at 1454442_at | 2043 | 0.654 | 0.0496 | No | ||
| 10 | NR2F1 | 1418157_at 1422412_x_at | 2215 | 0.608 | 0.0507 | No | ||
| 11 | KDELR3 | 1418538_at | 2235 | 0.603 | 0.0587 | No | ||
| 12 | GPR158 | 1438526_at 1459430_at | 2503 | 0.544 | 0.0544 | No | ||
| 13 | SCAMP5 | 1450247_a_at 1451224_at | 3406 | 0.398 | 0.0190 | No | ||
| 14 | FOSB | 1422134_at | 3800 | 0.354 | 0.0062 | No | ||
| 15 | SDHB | 1418005_at | 4204 | 0.320 | -0.0076 | No | ||
| 16 | GPC6 | 1419688_at 1419689_at 1428774_at 1437417_s_at 1440634_at 1441438_at 1443201_at 1445982_at | 4215 | 0.320 | -0.0033 | No | ||
| 17 | HAS1 | 1420590_at | 4248 | 0.317 | -0.0002 | No | ||
| 18 | WHSC1L1 | 1442085_at 1447931_at 1455668_at 1457630_at 1457793_a_at 1457794_at 1459907_a_at | 4433 | 0.305 | -0.0041 | No | ||
| 19 | MAP1LC3A | 1447883_x_at 1451290_at | 4557 | 0.297 | -0.0054 | No | ||
| 20 | SNF1LK | 1419766_at 1459601_at | 4595 | 0.294 | -0.0028 | No | ||
| 21 | CALM2 | 1422414_a_at 1423807_a_at | 4704 | 0.287 | -0.0035 | No | ||
| 22 | SMARCD1 | 1448913_at | 4844 | 0.279 | -0.0058 | No | ||
| 23 | SIX3 | 1426637_a_at 1426638_at | 5138 | 0.262 | -0.0153 | No | ||
| 24 | ALS2CR13 | 1434010_at 1444629_at 1446891_at | 5466 | 0.245 | -0.0267 | No | ||
| 25 | PCF11 | 1427159_at 1427160_at 1428724_at 1456489_at | 6484 | 0.201 | -0.0704 | No | ||
| 26 | PLCB3 | 1448661_at | 6579 | 0.197 | -0.0718 | No | ||
| 27 | GPM6B | 1423091_a_at 1425942_a_at 1453514_at | 6609 | 0.196 | -0.0702 | No | ||
| 28 | GPR3 | 1460275_at | 6994 | 0.183 | -0.0851 | No | ||
| 29 | SSTR3 | 1441603_at 1450577_at | 7377 | 0.169 | -0.1001 | No | ||
| 30 | EIF4A2 | 1450934_at | 7468 | 0.166 | -0.1018 | No | ||
| 31 | CLSTN3 | 1426989_at | 7609 | 0.162 | -0.1058 | No | ||
| 32 | PLK4 | 1419838_s_at 1426580_at 1452115_a_at | 7775 | 0.156 | -0.1111 | No | ||
| 33 | HOXB1 | 1453501_at | 8024 | 0.148 | -0.1203 | No | ||
| 34 | GTF2A1 | 1421357_at | 8084 | 0.146 | -0.1209 | No | ||
| 35 | PTPRU | 1416674_at 1446638_at | 8210 | 0.142 | -0.1245 | No | ||
| 36 | NOL4 | 1430189_at 1441447_at 1456387_at | 8449 | 0.135 | -0.1334 | No | ||
| 37 | TAF11 | 1446161_at 1451995_at | 8982 | 0.118 | -0.1561 | No | ||
| 38 | VGF | 1436094_at | 9063 | 0.115 | -0.1580 | No | ||
| 39 | RAB37 | 1433947_at 1450167_at | 9388 | 0.106 | -0.1713 | No | ||
| 40 | IRX4 | 1419539_at | 9850 | 0.092 | -0.1911 | No | ||
| 41 | SLC30A5 | 1422497_at | 10469 | 0.075 | -0.2183 | No | ||
| 42 | GATA1 | 1449232_at | 10714 | 0.067 | -0.2285 | No | ||
| 43 | CDK5R2 | 1450465_at | 10846 | 0.063 | -0.2336 | No | ||
| 44 | PEG3 | 1417355_at 1417356_at 1433924_at 1447731_at 1447732_x_at | 11083 | 0.057 | -0.2435 | No | ||
| 45 | GSPT1 | 1426736_at 1426737_at 1435000_at 1446550_at 1452168_x_at 1455173_at | 11318 | 0.051 | -0.2535 | No | ||
| 46 | DLX5 | 1449863_a_at | 11585 | 0.043 | -0.2651 | No | ||
| 47 | NTRK2 | 1420837_at 1420838_at 1435196_at 1435305_at 1437560_at 1446712_at | 11749 | 0.038 | -0.2720 | No | ||
| 48 | HOXA10 | 1431475_a_at 1446408_at | 12570 | 0.013 | -0.3093 | No | ||
| 49 | DLST | 1423710_at 1430161_at 1431011_at 1437775_at | 12963 | 0.001 | -0.3273 | No | ||
| 50 | MLF2 | 1423916_s_at | 13440 | -0.016 | -0.3489 | No | ||
| 51 | PPP2CB | 1421822_at 1421823_a_at 1431341_at | 13860 | -0.031 | -0.3676 | No | ||
| 52 | RPS27 | 1415716_a_at 1460699_at | 13992 | -0.035 | -0.3731 | No | ||
| 53 | CAMK2D | 1422659_at 1427763_a_at 1439168_at 1444031_at 1444281_at 1446663_at 1456844_at 1459457_at 1459600_at 1460630_at | 14224 | -0.045 | -0.3830 | No | ||
| 54 | TUBA2 | 1423846_x_at | 14560 | -0.057 | -0.3975 | No | ||
| 55 | OTUB2 | 1417575_at 1417576_a_at | 14589 | -0.058 | -0.3979 | No | ||
| 56 | G6PC | 1417880_at | 14682 | -0.062 | -0.4012 | No | ||
| 57 | ZIM2 | 1432297_at | 15060 | -0.078 | -0.4174 | No | ||
| 58 | VEGF | 1420909_at 1451959_a_at | 15178 | -0.085 | -0.4215 | No | ||
| 59 | BRAF | 1425693_at 1435434_at 1435480_at 1442749_at 1445786_at 1447940_a_at 1447941_x_at 1456505_at 1458641_at | 15601 | -0.105 | -0.4393 | No | ||
| 60 | HS6ST2 | 1420938_at 1420939_at 1446257_at 1450047_at | 15632 | -0.107 | -0.4391 | No | ||
| 61 | CD2AP | 1420906_at 1420907_at 1420908_at 1460140_at | 15765 | -0.114 | -0.4434 | No | ||
| 62 | ASXL1 | 1434561_at 1435077_at 1458380_at | 16231 | -0.144 | -0.4626 | No | ||
| 63 | GEM | 1426063_a_at | 16465 | -0.160 | -0.4710 | No | ||
| 64 | SMARCA5 | 1440048_at | 16594 | -0.170 | -0.4743 | No | ||
| 65 | CYLD | 1429617_at 1429618_at 1445213_at | 16799 | -0.187 | -0.4809 | No | ||
| 66 | ARMCX3 | 1424373_at 1460359_at | 16989 | -0.206 | -0.4866 | No | ||
| 67 | PAK1 | 1420979_at 1420980_at 1450070_s_at 1459029_at | 17237 | -0.231 | -0.4945 | No | ||
| 68 | XPR1 | 1426053_a_at 1426992_at 1426993_at 1437958_at 1444168_at 1444267_at 1450130_at 1452271_at | 17704 | -0.285 | -0.5116 | No | ||
| 69 | SFRS1 | 1428099_a_at 1428100_at 1430982_at 1434972_x_at 1452430_s_at 1453722_s_at 1457136_at | 17821 | -0.299 | -0.5126 | No | ||
| 70 | JMJD1B | 1428320_at 1438508_at 1458417_at | 17881 | -0.307 | -0.5108 | No | ||
| 71 | SLC25A5 | 1423772_x_at 1434801_x_at 1436874_x_at 1445944_at | 17914 | -0.312 | -0.5076 | No | ||
| 72 | MESDC1 | 1416397_at 1416398_at | 17934 | -0.316 | -0.5039 | No | ||
| 73 | MARCKS | 1415971_at 1415972_at 1430311_at 1437034_x_at 1456028_x_at | 18104 | -0.339 | -0.5066 | No | ||
| 74 | NINJ1 | 1438928_x_at 1441281_s_at 1447549_x_at 1448417_at | 18157 | -0.346 | -0.5039 | No | ||
| 75 | RING1 | 1422647_at | 18170 | -0.348 | -0.4993 | No | ||
| 76 | SOX10 | 1424985_a_at 1425369_a_at 1451689_a_at | 18434 | -0.384 | -0.5058 | No | ||
| 77 | OSBP | 1460350_at | 18551 | -0.404 | -0.5051 | No | ||
| 78 | ARRB2 | 1426239_s_at 1451987_at | 18580 | -0.409 | -0.5004 | No | ||
| 79 | HHIP | 1421426_at 1437933_at 1438083_at 1455277_at | 18641 | -0.421 | -0.4970 | No | ||
| 80 | FBXL11 | 1435329_at 1443336_at 1444358_at 1455942_at 1458449_at | 18689 | -0.432 | -0.4928 | No | ||
| 81 | CTCF | 1418330_at 1449042_at | 18744 | -0.444 | -0.4888 | No | ||
| 82 | MAFF | 1418936_at | 18877 | -0.470 | -0.4879 | No | ||
| 83 | BAHD1 | 1433703_s_at | 19954 | -0.763 | -0.5260 | Yes | ||
| 84 | TRIM39 | 1418851_at | 20024 | -0.790 | -0.5175 | Yes | ||
| 85 | PHF12 | 1434922_at 1453579_at | 20058 | -0.802 | -0.5073 | Yes | ||
| 86 | RFX5 | 1423103_at | 20107 | -0.820 | -0.4974 | Yes | ||
| 87 | FOS | 1423100_at | 20292 | -0.887 | -0.4928 | Yes | ||
| 88 | LMO4 | 1420981_a_at 1439651_at | 20457 | -0.961 | -0.4862 | Yes | ||
| 89 | VAMP2 | 1420833_at 1420834_at | 20497 | -0.982 | -0.4736 | Yes | ||
| 90 | TMEM1 | 1435430_at | 20778 | -1.168 | -0.4693 | Yes | ||
| 91 | FGF14 | 1435747_at 1447221_x_at 1447480_at 1449958_a_at 1458830_at 1459073_x_at | 20785 | -1.172 | -0.4524 | Yes | ||
| 92 | MBNL2 | 1433754_at 1436858_at 1446475_at 1455827_at 1457237_at 1457477_at | 21048 | -1.411 | -0.4436 | Yes | ||
| 93 | ARIH1 | 1427188_at 1427189_at 1436498_at 1441022_at 1459397_at 1460090_at | 21144 | -1.526 | -0.4256 | Yes | ||
| 94 | USP14 | 1416208_at 1437714_x_at 1439201_at 1455829_at | 21231 | -1.636 | -0.4055 | Yes | ||
| 95 | KLF13 | 1432543_a_at 1456030_at | 21412 | -1.910 | -0.3857 | Yes | ||
| 96 | NLK | 1419112_at 1435970_at 1443279_at 1445427_at 1456467_s_at | 21448 | -1.981 | -0.3582 | Yes | ||
| 97 | DUSP1 | 1448830_at | 21513 | -2.123 | -0.3300 | Yes | ||
| 98 | PAFAH1B1 | 1417086_at 1427703_at 1439656_at 1441404_at 1444125_at 1448578_at 1456947_at 1460199_a_at | 21667 | -2.566 | -0.2993 | Yes | ||
| 99 | CDC42 | 1415724_a_at 1435807_at 1449574_a_at 1460708_s_at | 21696 | -2.726 | -0.2606 | Yes | ||
| 100 | CCND2 | 1416122_at 1416123_at 1416124_at 1430127_a_at 1434745_at 1448229_s_at 1455956_x_at | 21702 | -2.755 | -0.2203 | Yes | ||
| 101 | LUC7L2 | 1423251_at 1427087_at 1436165_at 1436766_at 1436767_at 1437761_at 1458423_at | 21834 | -3.435 | -0.1759 | Yes | ||
| 102 | CDKN1B | 1419497_at 1434045_at | 21843 | -3.508 | -0.1248 | Yes | ||
| 103 | PNRC1 | 1433668_at 1438524_x_at 1439513_at | 21934 | -8.778 | 0.0000 | Yes |