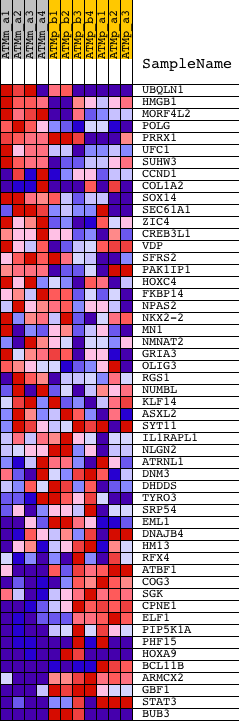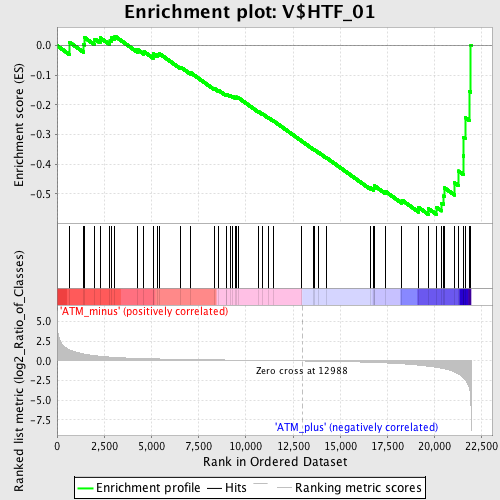
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | V$HTF_01 |
| Enrichment Score (ES) | -0.56948864 |
| Normalized Enrichment Score (NES) | -1.582712 |
| Nominal p-value | 0.008431703 |
| FDR q-value | 0.06326536 |
| FWER p-Value | 0.608 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | UBQLN1 | 1419385_a_at 1424368_s_at 1450345_at | 650 | 1.414 | 0.0120 | No | ||
| 2 | HMGB1 | 1416176_at 1425048_a_at 1435324_x_at | 1401 | 0.890 | 0.0040 | No | ||
| 3 | MORF4L2 | 1415778_at 1439413_x_at | 1447 | 0.869 | 0.0275 | No | ||
| 4 | POLG | 1423272_at 1423273_at 1437564_at | 1998 | 0.671 | 0.0222 | No | ||
| 5 | PRRX1 | 1425526_a_at 1425527_at 1425528_at 1432129_a_at 1439774_at 1458004_at | 2285 | 0.593 | 0.0266 | No | ||
| 6 | UFC1 | 1416327_at | 2799 | 0.482 | 0.0174 | No | ||
| 7 | SUHW3 | 1427983_at | 2895 | 0.465 | 0.0267 | No | ||
| 8 | CCND1 | 1417419_at 1417420_at 1448698_at | 3060 | 0.439 | 0.0322 | No | ||
| 9 | COL1A2 | 1446326_at 1450857_a_at | 4254 | 0.317 | -0.0130 | No | ||
| 10 | SOX14 | 1427609_at | 4599 | 0.294 | -0.0200 | No | ||
| 11 | SEC61A1 | 1416189_a_at 1416190_a_at 1416191_at 1434986_a_at 1448242_at 1455987_at | 5087 | 0.265 | -0.0345 | No | ||
| 12 | ZIC4 | 1421539_at 1456417_at | 5095 | 0.264 | -0.0270 | No | ||
| 13 | CREB3L1 | 1419295_at | 5306 | 0.254 | -0.0291 | No | ||
| 14 | VDP | 1424274_at | 5411 | 0.249 | -0.0265 | No | ||
| 15 | SFRS2 | 1415807_s_at 1427504_s_at 1427816_at 1452439_s_at | 6535 | 0.199 | -0.0720 | No | ||
| 16 | PAK1IP1 | 1423766_at 1430875_a_at | 7069 | 0.180 | -0.0910 | No | ||
| 17 | HOXC4 | 1422870_at | 8342 | 0.138 | -0.1451 | No | ||
| 18 | FKBP14 | 1433481_at 1458225_at | 8570 | 0.131 | -0.1516 | No | ||
| 19 | NPAS2 | 1421036_at 1421037_at | 8952 | 0.119 | -0.1655 | No | ||
| 20 | NKX2-2 | 1421112_at | 8992 | 0.117 | -0.1638 | No | ||
| 21 | MN1 | 1454867_at | 9165 | 0.112 | -0.1683 | No | ||
| 22 | NMNAT2 | 1436155_at | 9297 | 0.109 | -0.1711 | No | ||
| 23 | GRIA3 | 1420563_at 1434728_at | 9471 | 0.104 | -0.1760 | No | ||
| 24 | OLIG3 | 1421728_at | 9474 | 0.103 | -0.1730 | No | ||
| 25 | RGS1 | 1417601_at | 9606 | 0.099 | -0.1761 | No | ||
| 26 | NUMBL | 1416491_at | 10685 | 0.068 | -0.2233 | No | ||
| 27 | KLF14 | 1457397_at | 10877 | 0.062 | -0.2302 | No | ||
| 28 | ASXL2 | 1439063_at 1444754_at 1453295_at 1458366_at 1460597_at | 11210 | 0.054 | -0.2438 | No | ||
| 29 | SYT11 | 1429314_at 1429315_at 1429729_at 1436176_at 1449264_at 1455176_a_at | 11458 | 0.047 | -0.2537 | No | ||
| 30 | IL1RAPL1 | 1445821_at 1458964_at | 12940 | 0.002 | -0.3213 | No | ||
| 31 | NLGN2 | 1455143_at 1460013_at | 13586 | -0.020 | -0.3502 | No | ||
| 32 | ATRNL1 | 1419922_s_at 1446401_at 1449666_at 1460152_at | 13618 | -0.022 | -0.3510 | No | ||
| 33 | DNM3 | 1425403_at 1436624_at 1436875_at 1438801_at 1445540_at 1446265_at 1446431_at 1446694_at 1457405_at | 13865 | -0.031 | -0.3613 | No | ||
| 34 | DHDDS | 1422548_at 1425520_a_at 1450654_a_at | 14270 | -0.046 | -0.3784 | No | ||
| 35 | TYRO3 | 1425248_a_at 1425249_a_at | 16578 | -0.169 | -0.4789 | No | ||
| 36 | SRP54 | 1430476_at 1441233_at 1441480_at | 16776 | -0.185 | -0.4824 | No | ||
| 37 | EML1 | 1458317_at | 16797 | -0.187 | -0.4778 | No | ||
| 38 | DNAJB4 | 1431734_a_at 1439524_at 1443611_at 1451177_at | 16805 | -0.187 | -0.4726 | No | ||
| 39 | HM13 | 1417287_at 1428855_at 1428856_at 1438456_at 1438865_at 1455631_at | 17398 | -0.249 | -0.4923 | No | ||
| 40 | RFX4 | 1432053_at 1436931_at 1443312_at | 18263 | -0.362 | -0.5211 | No | ||
| 41 | ATBF1 | 1420649_at 1420650_at 1429725_at 1445941_at 1446094_at 1449947_s_at 1453267_at 1460006_at | 19148 | -0.532 | -0.5458 | No | ||
| 42 | COG3 | 1434654_at 1456286_at | 19667 | -0.666 | -0.5499 | Yes | ||
| 43 | SGK | 1416041_at | 20087 | -0.813 | -0.5450 | Yes | ||
| 44 | CPNE1 | 1433439_at | 20367 | -0.916 | -0.5308 | Yes | ||
| 45 | ELF1 | 1417540_at 1439319_at | 20470 | -0.965 | -0.5069 | Yes | ||
| 46 | PIP5K1A | 1421833_at 1421834_at 1450389_s_at | 20517 | -0.993 | -0.4798 | Yes | ||
| 47 | PHF15 | 1430081_at 1455345_at | 21049 | -1.412 | -0.4624 | Yes | ||
| 48 | HOXA9 | 1421579_at 1455626_at | 21257 | -1.668 | -0.4227 | Yes | ||
| 49 | BCL11B | 1440137_at 1441762_at 1450339_a_at | 21512 | -2.120 | -0.3718 | Yes | ||
| 50 | ARMCX2 | 1415780_a_at 1437849_x_at 1456739_x_at | 21535 | -2.165 | -0.3089 | Yes | ||
| 51 | GBF1 | 1415709_s_at 1415757_at 1432677_at 1438207_at | 21611 | -2.381 | -0.2421 | Yes | ||
| 52 | STAT3 | 1424272_at 1426587_a_at 1459961_a_at 1460700_at | 21823 | -3.342 | -0.1532 | Yes | ||
| 53 | BUB3 | 1416815_s_at 1448058_s_at 1448473_at 1459104_at 1459918_at | 21913 | -5.367 | 0.0010 | Yes |