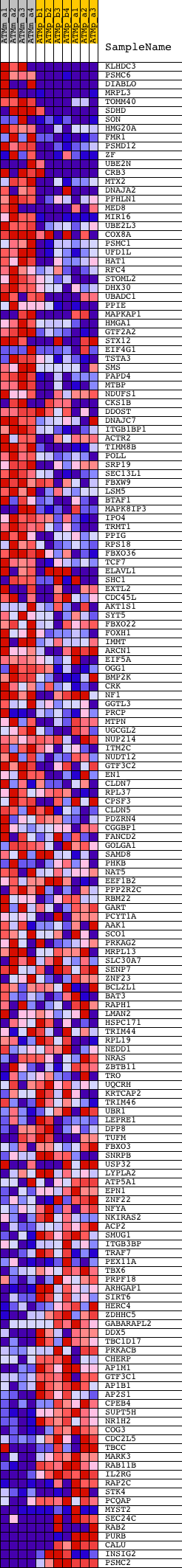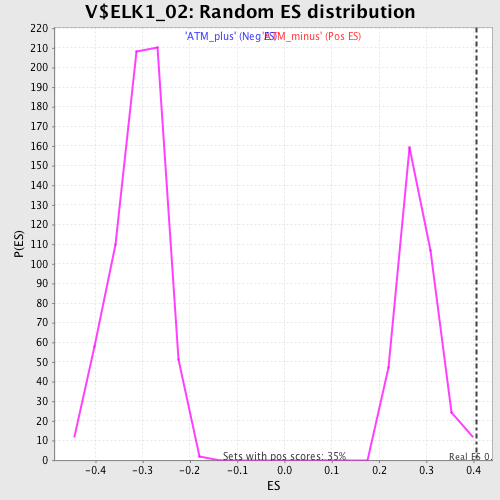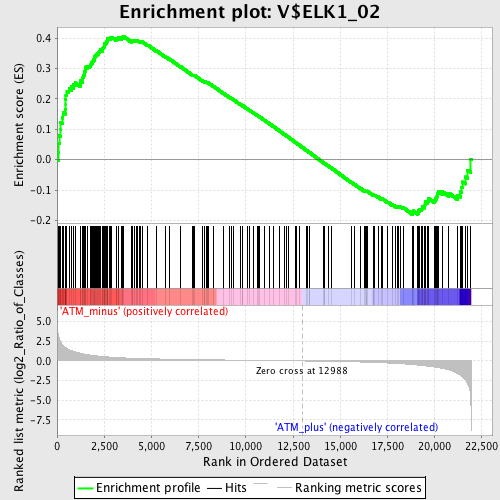
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | V$ELK1_02 |
| Enrichment Score (ES) | 0.40630597 |
| Normalized Enrichment Score (NES) | 1.4414225 |
| Nominal p-value | 0.0028653296 |
| FDR q-value | 0.37538525 |
| FWER p-Value | 0.924 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | KLHDC3 | 1415991_a_at 1448164_at 1454747_a_at 1455910_at | 83 | 3.057 | 0.0252 | Yes | ||
| 2 | PSMC6 | 1417771_a_at | 90 | 3.001 | 0.0533 | Yes | ||
| 3 | DIABLO | 1417683_at 1425768_at | 115 | 2.808 | 0.0789 | Yes | ||
| 4 | MRPL3 | 1422463_a_at 1422464_at 1431006_at 1440636_at | 179 | 2.479 | 0.0995 | Yes | ||
| 5 | TOMM40 | 1426118_a_at 1459680_at | 184 | 2.454 | 0.1225 | Yes | ||
| 6 | SDHD | 1428235_at 1437489_x_at | 273 | 2.115 | 0.1385 | Yes | ||
| 7 | SON | 1420951_a_at 1420952_at 1435862_at 1437924_at 1438871_at 1439074_a_at 1455634_at 1456630_x_at 1457596_at | 311 | 1.958 | 0.1554 | Yes | ||
| 8 | HMG20A | 1416722_at 1428920_at 1442683_at 1442684_x_at | 429 | 1.716 | 0.1663 | Yes | ||
| 9 | FMR1 | 1423369_at 1426086_a_at 1452550_a_at | 448 | 1.689 | 0.1815 | Yes | ||
| 10 | PSMD12 | 1448492_a_at | 450 | 1.687 | 0.1974 | Yes | ||
| 11 | ZF | 1425698_a_at 1438302_at 1452856_at 1452857_at | 466 | 1.660 | 0.2125 | Yes | ||
| 12 | UBE2N | 1422559_at 1435384_at | 522 | 1.575 | 0.2249 | Yes | ||
| 13 | CRB3 | 1424809_at 1434851_s_at 1455115_a_at | 629 | 1.436 | 0.2336 | Yes | ||
| 14 | MTX2 | 1430500_s_at | 768 | 1.295 | 0.2395 | Yes | ||
| 15 | DNAJA2 | 1417182_at 1417183_at 1429476_s_at 1457233_at | 867 | 1.211 | 0.2465 | Yes | ||
| 16 | PPHLN1 | 1434822_at 1435766_at | 957 | 1.143 | 0.2533 | Yes | ||
| 17 | MED8 | 1431423_a_at 1442417_at | 1220 | 0.990 | 0.2506 | Yes | ||
| 18 | MIR16 | 1418444_a_at | 1236 | 0.981 | 0.2592 | Yes | ||
| 19 | UBE2L3 | 1417907_at 1417908_s_at 1448879_at 1448880_at | 1357 | 0.918 | 0.2624 | Yes | ||
| 20 | COX8A | 1416112_at 1436409_at 1448222_x_at | 1367 | 0.911 | 0.2706 | Yes | ||
| 21 | PSMC1 | 1416005_at | 1384 | 0.901 | 0.2784 | Yes | ||
| 22 | UFD1L | 1418087_at 1432367_a_at | 1435 | 0.873 | 0.2844 | Yes | ||
| 23 | HAT1 | 1428061_at | 1458 | 0.864 | 0.2916 | Yes | ||
| 24 | RFC4 | 1424321_at 1438161_s_at | 1513 | 0.845 | 0.2971 | Yes | ||
| 25 | STOML2 | 1448774_at | 1521 | 0.842 | 0.3048 | Yes | ||
| 26 | DHX30 | 1416140_a_at 1438832_x_at 1438969_x_at 1453251_at | 1624 | 0.800 | 0.3077 | Yes | ||
| 27 | UBADC1 | 1416963_at 1446898_at | 1741 | 0.755 | 0.3095 | Yes | ||
| 28 | PPIE | 1450657_at | 1791 | 0.735 | 0.3142 | Yes | ||
| 29 | MAPKAP1 | 1417284_at 1427212_at 1442692_at | 1804 | 0.731 | 0.3206 | Yes | ||
| 30 | HMGA1 | 1416184_s_at | 1893 | 0.703 | 0.3232 | Yes | ||
| 31 | GTF2A2 | 1433528_at | 1919 | 0.694 | 0.3287 | Yes | ||
| 32 | STX12 | 1435982_at 1448936_at | 1977 | 0.677 | 0.3325 | Yes | ||
| 33 | EIF4G1 | 1427036_a_at 1427037_at 1438686_at | 1995 | 0.672 | 0.3381 | Yes | ||
| 34 | TSTA3 | 1448495_at | 2009 | 0.666 | 0.3438 | Yes | ||
| 35 | SMS | 1428699_at 1434190_at | 2072 | 0.647 | 0.3471 | Yes | ||
| 36 | PAPD4 | 1419183_at 1433568_at | 2137 | 0.631 | 0.3501 | Yes | ||
| 37 | MTBP | 1448638_at | 2208 | 0.610 | 0.3527 | Yes | ||
| 38 | NDUFS1 | 1425143_a_at 1453354_at | 2233 | 0.603 | 0.3573 | Yes | ||
| 39 | CKS1B | 1416698_a_at 1439514_at 1448441_at 1456268_at | 2273 | 0.595 | 0.3611 | Yes | ||
| 40 | DDOST | 1416493_at 1420179_at | 2383 | 0.572 | 0.3615 | Yes | ||
| 41 | DNAJC7 | 1448362_at 1459704_at | 2393 | 0.570 | 0.3665 | Yes | ||
| 42 | ITGB1BP1 | 1422157_a_at 1445697_at | 2439 | 0.561 | 0.3698 | Yes | ||
| 43 | ACTR2 | 1441660_at 1452587_at | 2486 | 0.547 | 0.3729 | Yes | ||
| 44 | TIMM8B | 1431665_a_at | 2501 | 0.544 | 0.3774 | Yes | ||
| 45 | POLL | 1449352_at | 2516 | 0.540 | 0.3818 | Yes | ||
| 46 | SRP19 | 1450891_at | 2586 | 0.528 | 0.3837 | Yes | ||
| 47 | SEC13L1 | 1416241_at | 2617 | 0.521 | 0.3872 | Yes | ||
| 48 | FBXW9 | 1425857_at 1428120_at | 2642 | 0.515 | 0.3910 | Yes | ||
| 49 | LSM5 | 1418656_at | 2669 | 0.511 | 0.3947 | Yes | ||
| 50 | BTAF1 | 1435248_a_at 1435249_at 1435953_at | 2681 | 0.507 | 0.3990 | Yes | ||
| 51 | MAPK8IP3 | 1416437_a_at 1425975_a_at 1436676_at 1460628_at | 2765 | 0.489 | 0.3998 | Yes | ||
| 52 | IPO4 | 1436420_a_at 1450814_a_at | 2849 | 0.472 | 0.4005 | Yes | ||
| 53 | TRMT1 | 1436494_x_at 1437314_a_at 1451249_at | 2903 | 0.464 | 0.4024 | Yes | ||
| 54 | PPIG | 1434475_at 1435341_at 1436505_at | 3123 | 0.431 | 0.3965 | Yes | ||
| 55 | RPS18 | 1421837_at 1421838_at 1435712_a_at 1448739_x_at 1450390_x_at 1455572_x_at | 3150 | 0.427 | 0.3993 | Yes | ||
| 56 | FBXO36 | 1419305_a_at 1449418_s_at | 3228 | 0.420 | 0.3997 | Yes | ||
| 57 | TCF7 | 1433471_at 1450461_at | 3248 | 0.417 | 0.4028 | Yes | ||
| 58 | ELAVL1 | 1428653_x_at 1431037_a_at 1440373_at 1440464_at 1445521_at 1447109_at 1448151_at 1452858_at | 3415 | 0.396 | 0.3990 | Yes | ||
| 59 | SHC1 | 1422853_at 1422854_at | 3459 | 0.392 | 0.4007 | Yes | ||
| 60 | EXTL2 | 1422538_at 1422539_at | 3481 | 0.389 | 0.4034 | Yes | ||
| 61 | CDC45L | 1416575_at 1457838_at | 3499 | 0.388 | 0.4063 | Yes | ||
| 62 | AKT1S1 | 1428158_at 1437736_at 1452684_at | 3927 | 0.343 | 0.3899 | No | ||
| 63 | SYT5 | 1422531_at | 3988 | 0.338 | 0.3904 | No | ||
| 64 | FBXO22 | 1426592_a_at 1426593_a_at 1452121_at | 3996 | 0.337 | 0.3933 | No | ||
| 65 | FOXH1 | 1422212_at 1422213_s_at 1437779_at | 4091 | 0.329 | 0.3921 | No | ||
| 66 | IMMT | 1429533_at 1429534_a_at 1433470_a_at 1437126_at | 4185 | 0.322 | 0.3908 | No | ||
| 67 | ARCN1 | 1423743_at 1436062_at 1444372_at 1451089_a_at | 4240 | 0.318 | 0.3914 | No | ||
| 68 | EIF5A | 1437859_x_at 1451470_s_at | 4360 | 0.310 | 0.3888 | No | ||
| 69 | OGG1 | 1430078_a_at 1448815_at | 4402 | 0.307 | 0.3899 | No | ||
| 70 | BMP2K | 1421103_at 1437419_at 1458370_at | 4515 | 0.299 | 0.3876 | No | ||
| 71 | CRK | 1416201_at 1425855_a_at 1436835_at 1448248_at 1460176_at | 4804 | 0.281 | 0.3770 | No | ||
| 72 | NF1 | 1438067_at 1441523_at 1443097_at 1452525_a_at | 5271 | 0.256 | 0.3580 | No | ||
| 73 | GGTL3 | 1451094_at | 5729 | 0.232 | 0.3392 | No | ||
| 74 | PRCP | 1431361_at 1444419_at 1452190_at 1452191_at | 5951 | 0.222 | 0.3312 | No | ||
| 75 | MTPN | 1420472_at 1420473_at 1420474_at 1420475_at 1437457_a_at 1459597_at | 6551 | 0.199 | 0.3055 | No | ||
| 76 | UGCGL2 | 1429836_at 1431602_a_at | 7180 | 0.176 | 0.2783 | No | ||
| 77 | NUP214 | 1434351_at 1456503_at 1457815_at | 7240 | 0.174 | 0.2773 | No | ||
| 78 | ITM2C | 1415961_at | 7286 | 0.173 | 0.2768 | No | ||
| 79 | NUDT12 | 1453139_at | 7699 | 0.159 | 0.2594 | No | ||
| 80 | GTF3C2 | 1428448_a_at 1428449_at 1440307_at 1452780_at 1452781_a_at | 7805 | 0.155 | 0.2561 | No | ||
| 81 | EN1 | 1418618_at | 7825 | 0.155 | 0.2567 | No | ||
| 82 | CLDN7 | 1448393_at | 7901 | 0.152 | 0.2547 | No | ||
| 83 | RPL37 | 1453729_a_at | 7946 | 0.150 | 0.2541 | No | ||
| 84 | CPSF3 | 1422855_at 1437328_x_at 1437852_x_at | 7992 | 0.149 | 0.2534 | No | ||
| 85 | CLDN5 | 1417839_at | 8305 | 0.139 | 0.2404 | No | ||
| 86 | PDZRN4 | 1456512_at | 8792 | 0.124 | 0.2192 | No | ||
| 87 | CGGBP1 | 1454641_at 1455658_at | 9132 | 0.113 | 0.2047 | No | ||
| 88 | FANCD2 | 1430916_at 1438339_at 1439091_at 1442092_at | 9224 | 0.111 | 0.2016 | No | ||
| 89 | GOLGA1 | 1431120_a_at 1432054_at 1435027_at 1446087_at | 9328 | 0.108 | 0.1979 | No | ||
| 90 | SAMD8 | 1434402_at 1453730_at 1458408_at | 9699 | 0.096 | 0.1818 | No | ||
| 91 | PHKB | 1434511_at 1442053_at | 9792 | 0.094 | 0.1785 | No | ||
| 92 | NAT5 | 1418244_at 1422961_at | 9839 | 0.093 | 0.1772 | No | ||
| 93 | EEF1B2 | 1448252_a_at | 10092 | 0.085 | 0.1665 | No | ||
| 94 | PPP2R2C | 1438054_x_at 1438671_at 1443804_at | 10213 | 0.082 | 0.1617 | No | ||
| 95 | RBM22 | 1452675_at | 10412 | 0.076 | 0.1533 | No | ||
| 96 | GART | 1416283_at 1424435_a_at 1424436_at 1441911_x_at | 10617 | 0.070 | 0.1446 | No | ||
| 97 | PCYT1A | 1421957_a_at 1424453_at 1438011_at 1450434_s_at | 10666 | 0.069 | 0.1431 | No | ||
| 98 | AAK1 | 1420026_at 1435038_s_at 1441782_at | 10729 | 0.067 | 0.1409 | No | ||
| 99 | SCO1 | 1430300_at | 11005 | 0.059 | 0.1288 | No | ||
| 100 | PRKAG2 | 1423831_at 1423832_at 1431263_at 1451140_s_at 1457885_at | 11273 | 0.052 | 0.1170 | No | ||
| 101 | MRPL13 | 1424204_at 1435966_x_at 1459856_at 1460354_a_at | 11468 | 0.046 | 0.1085 | No | ||
| 102 | SLC30A7 | 1450697_at | 11762 | 0.038 | 0.0954 | No | ||
| 103 | SENP7 | 1430974_a_at 1436860_at 1438413_at | 12025 | 0.029 | 0.0837 | No | ||
| 104 | ZNF23 | 1427104_at 1456824_at 1459066_at | 12137 | 0.026 | 0.0788 | No | ||
| 105 | BCL2L1 | 1420887_a_at 1420888_at 1426050_at 1426191_a_at | 12279 | 0.022 | 0.0726 | No | ||
| 106 | BAT3 | 1433890_a_at | 12600 | 0.012 | 0.0580 | No | ||
| 107 | RAPH1 | 1434302_at 1434303_at 1444586_at 1456918_at | 12678 | 0.010 | 0.0545 | No | ||
| 108 | LMAN2 | 1423074_at 1423075_at | 12840 | 0.005 | 0.0472 | No | ||
| 109 | HSPC171 | 1419287_at | 13218 | -0.008 | 0.0299 | No | ||
| 110 | TRIM44 | 1421869_at 1421870_at 1431932_s_at 1440013_at | 13257 | -0.009 | 0.0283 | No | ||
| 111 | RPL19 | 1416219_at | 13348 | -0.012 | 0.0243 | No | ||
| 112 | NEDD1 | 1423196_at 1450899_at | 13350 | -0.012 | 0.0243 | No | ||
| 113 | NRAS | 1422688_a_at 1454060_a_at | 14084 | -0.039 | -0.0090 | No | ||
| 114 | ZBTB11 | 1454826_at | 14150 | -0.042 | -0.0116 | No | ||
| 115 | TRO | 1421201_a_at 1426016_a_at | 14175 | -0.043 | -0.0123 | No | ||
| 116 | UQCRH | 1452133_at 1453229_s_at | 14394 | -0.051 | -0.0218 | No | ||
| 117 | KRTCAP2 | 1417058_a_at 1417059_at | 14544 | -0.056 | -0.0281 | No | ||
| 118 | TRIM46 | 1460568_at | 15583 | -0.104 | -0.0748 | No | ||
| 119 | UBR1 | 1420971_at 1440552_at 1446438_at 1450066_at 1457796_at 1458033_at | 15732 | -0.112 | -0.0806 | No | ||
| 120 | LEPRE1 | 1421462_a_at 1452752_at | 16082 | -0.133 | -0.0953 | No | ||
| 121 | DPP8 | 1428697_at 1431064_at 1445705_x_at | 16255 | -0.145 | -0.1019 | No | ||
| 122 | TUFM | 1433952_at 1449673_s_at | 16341 | -0.152 | -0.1043 | No | ||
| 123 | FBXO3 | 1423490_at 1423491_at 1431078_at 1441019_at 1455047_at | 16381 | -0.154 | -0.1046 | No | ||
| 124 | SNRPB | 1419260_a_at 1437193_s_at | 16391 | -0.155 | -0.1036 | No | ||
| 125 | USP32 | 1430403_at 1436159_at 1439857_at 1440399_at 1443689_at 1459857_at | 16411 | -0.156 | -0.1030 | No | ||
| 126 | LYPLA2 | 1417433_at | 16730 | -0.181 | -0.1159 | No | ||
| 127 | ATP5A1 | 1420037_at 1423111_at 1449710_s_at | 16773 | -0.185 | -0.1161 | No | ||
| 128 | EPN1 | 1427039_at | 16825 | -0.190 | -0.1166 | No | ||
| 129 | ZNF22 | 1451244_a_at | 16995 | -0.206 | -0.1224 | No | ||
| 130 | NFYA | 1422082_a_at 1427808_at 1452560_a_at | 17163 | -0.224 | -0.1280 | No | ||
| 131 | NKIRAS2 | 1424416_at | 17226 | -0.230 | -0.1286 | No | ||
| 132 | ACP2 | 1424654_at 1424655_at | 17518 | -0.262 | -0.1395 | No | ||
| 133 | SMUG1 | 1428763_at | 17753 | -0.291 | -0.1475 | No | ||
| 134 | ITGB3BP | 1420665_at 1442993_at 1449953_at 1456097_a_at | 17943 | -0.318 | -0.1532 | No | ||
| 135 | TRAF7 | 1424320_a_at | 18034 | -0.328 | -0.1542 | No | ||
| 136 | PEX11A | 1419365_at 1449442_at | 18091 | -0.337 | -0.1536 | No | ||
| 137 | TBX6 | 1449868_at | 18194 | -0.351 | -0.1549 | No | ||
| 138 | PRPF18 | 1429614_at | 18323 | -0.370 | -0.1573 | No | ||
| 139 | ARHGAP1 | 1424307_at 1451309_at | 18801 | -0.456 | -0.1749 | No | ||
| 140 | SIRT6 | 1429631_at | 18831 | -0.460 | -0.1719 | No | ||
| 141 | HERC4 | 1427032_at 1431716_at 1456677_at | 18855 | -0.464 | -0.1685 | No | ||
| 142 | ZDHHC5 | 1426310_at 1426311_s_at | 19110 | -0.522 | -0.1753 | No | ||
| 143 | GABARAPL2 | 1423187_at 1441376_at 1444956_at 1459477_at | 19131 | -0.528 | -0.1712 | No | ||
| 144 | DDX5 | 1419653_a_at 1423645_a_at 1433809_at 1433810_x_at 1438371_x_at 1454793_x_at | 19138 | -0.529 | -0.1664 | No | ||
| 145 | TBC1D17 | 1423659_a_at | 19201 | -0.545 | -0.1641 | No | ||
| 146 | PRKACB | 1420610_at 1420611_at 1457197_at | 19297 | -0.572 | -0.1631 | No | ||
| 147 | CHERP | 1431770_at 1436885_a_at 1451896_a_at | 19326 | -0.580 | -0.1588 | No | ||
| 148 | AP1M1 | 1416307_at 1459832_s_at | 19345 | -0.583 | -0.1541 | No | ||
| 149 | GTF3C1 | 1434066_at 1438425_at 1456024_at 1459673_at | 19452 | -0.608 | -0.1533 | No | ||
| 150 | AP1B1 | 1416279_at | 19482 | -0.615 | -0.1488 | No | ||
| 151 | AP2S1 | 1433612_at | 19489 | -0.616 | -0.1432 | No | ||
| 152 | CPEB4 | 1420617_at 1420618_at 1449931_at | 19505 | -0.620 | -0.1380 | No | ||
| 153 | SUPT5H | 1424255_at | 19633 | -0.656 | -0.1376 | No | ||
| 154 | NR1H2 | 1416353_at | 19666 | -0.666 | -0.1328 | No | ||
| 155 | COG3 | 1434654_at 1456286_at | 19667 | -0.666 | -0.1265 | No | ||
| 156 | CDC2L5 | 1440833_at 1452897_at 1458313_at | 19963 | -0.766 | -0.1328 | No | ||
| 157 | TBCC | 1424644_at 1451423_at | 20034 | -0.792 | -0.1285 | No | ||
| 158 | MARK3 | 1418316_a_at | 20071 | -0.808 | -0.1225 | No | ||
| 159 | RAB11B | 1423448_at | 20120 | -0.824 | -0.1169 | No | ||
| 160 | IL2RG | 1416295_a_at 1416296_at | 20146 | -0.833 | -0.1101 | No | ||
| 161 | RAP2C | 1428119_a_at 1437016_x_at 1460430_at | 20187 | -0.845 | -0.1040 | No | ||
| 162 | STK4 | 1421107_at 1434159_at 1436015_s_at | 20396 | -0.931 | -0.1047 | No | ||
| 163 | PCQAP | 1441617_at 1444680_at 1444688_at 1448435_at | 20746 | -1.146 | -0.1099 | No | ||
| 164 | MYST2 | 1433433_at 1440746_at 1447631_at | 21223 | -1.619 | -0.1164 | No | ||
| 165 | SEC24C | 1430250_at 1452147_at | 21365 | -1.826 | -0.1056 | No | ||
| 166 | RAB2 | 1418621_at 1418622_at 1418623_at 1419945_s_at 1419946_s_at 1436477_x_at 1449676_at | 21428 | -1.934 | -0.0901 | No | ||
| 167 | PURB | 1419640_at 1419641_at 1419642_at 1428254_at 1436940_at | 21489 | -2.064 | -0.0733 | No | ||
| 168 | CALU | 1415870_at 1441477_at | 21637 | -2.443 | -0.0569 | No | ||
| 169 | INSIG2 | 1417980_a_at 1417981_at 1417982_at | 21744 | -2.907 | -0.0342 | No | ||
| 170 | PSMC2 | 1426611_at 1435859_x_at 1443307_at | 21900 | -4.520 | 0.0016 | No |