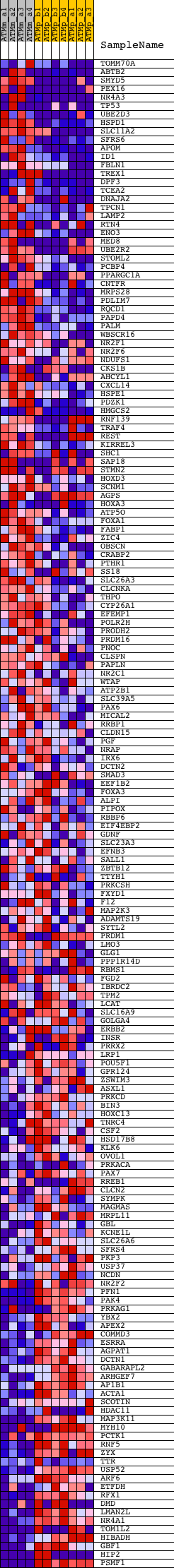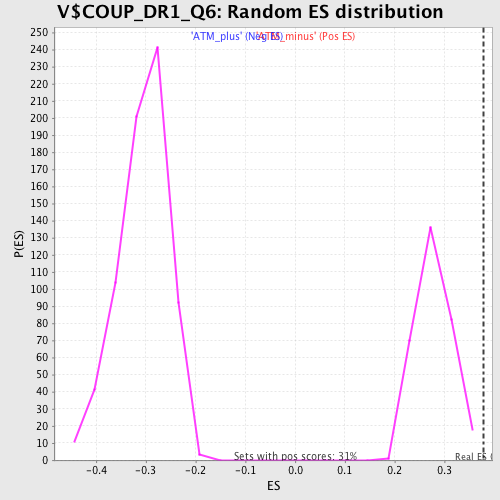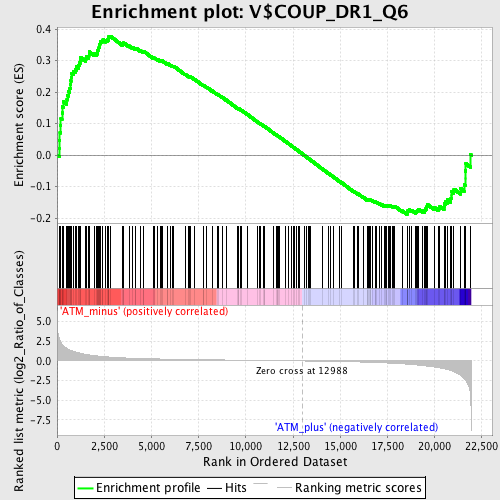
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | V$COUP_DR1_Q6 |
| Enrichment Score (ES) | 0.37809002 |
| Normalized Enrichment Score (NES) | 1.3596807 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.27021876 |
| FWER p-Value | 0.997 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TOMM70A | 1417192_at 1426675_at 1426676_s_at 1447239_at 1457353_at | 118 | 2.784 | 0.0215 | Yes | ||
| 2 | ABTB2 | 1433453_a_at 1433454_at 1443497_at 1444771_at | 127 | 2.720 | 0.0475 | Yes | ||
| 3 | SMYD5 | 1425479_at 1447749_at 1451722_s_at | 150 | 2.594 | 0.0716 | Yes | ||
| 4 | PEX16 | 1425021_a_at 1436253_at 1447801_x_at | 182 | 2.464 | 0.0941 | Yes | ||
| 5 | NR4A3 | 1421079_at 1421080_at | 201 | 2.364 | 0.1162 | Yes | ||
| 6 | TP53 | 1426538_a_at 1427739_a_at 1438808_at 1457623_x_at 1459780_at 1459781_x_at | 262 | 2.149 | 0.1342 | Yes | ||
| 7 | UBE2D3 | 1423112_at 1423113_a_at 1423114_at 1450858_a_at 1450859_s_at 1455479_a_at 1455480_s_at | 276 | 2.104 | 0.1540 | Yes | ||
| 8 | HSPD1 | 1426351_at | 323 | 1.927 | 0.1705 | Yes | ||
| 9 | SLC11A2 | 1417584_at 1426441_at 1441709_at 1447333_at 1452078_a_at | 519 | 1.576 | 0.1769 | Yes | ||
| 10 | SFRS6 | 1416720_at 1416721_s_at 1447898_s_at 1448454_at | 557 | 1.530 | 0.1900 | Yes | ||
| 11 | APOM | 1419095_a_at 1419096_at | 609 | 1.458 | 0.2018 | Yes | ||
| 12 | ID1 | 1425895_a_at | 674 | 1.389 | 0.2123 | Yes | ||
| 13 | FBLN1 | 1422540_at 1439688_at 1451119_a_at | 703 | 1.357 | 0.2241 | Yes | ||
| 14 | TREX1 | 1450672_a_at 1459618_at | 728 | 1.332 | 0.2359 | Yes | ||
| 15 | DPF3 | 1441659_at 1442686_at 1450162_at | 740 | 1.317 | 0.2482 | Yes | ||
| 16 | TCEA2 | 1418107_at 1440791_x_at | 776 | 1.291 | 0.2591 | Yes | ||
| 17 | DNAJA2 | 1417182_at 1417183_at 1429476_s_at 1457233_at | 867 | 1.211 | 0.2667 | Yes | ||
| 18 | TPCN1 | 1434930_at 1451771_at 1451772_at | 959 | 1.141 | 0.2735 | Yes | ||
| 19 | LAMP2 | 1416343_a_at 1416344_at 1428094_at 1434503_s_at | 1034 | 1.096 | 0.2808 | Yes | ||
| 20 | RTN4 | 1421116_a_at 1435284_at 1437224_at 1439650_at 1442760_x_at 1452649_at | 1143 | 1.033 | 0.2858 | Yes | ||
| 21 | ENO3 | 1417951_at 1449631_at | 1197 | 1.005 | 0.2931 | Yes | ||
| 22 | MED8 | 1431423_a_at 1442417_at | 1220 | 0.990 | 0.3017 | Yes | ||
| 23 | UBE2R2 | 1415768_a_at 1443742_x_at 1444460_at | 1231 | 0.985 | 0.3108 | Yes | ||
| 24 | STOML2 | 1448774_at | 1521 | 0.842 | 0.3056 | Yes | ||
| 25 | PCBP4 | 1433658_x_at 1449054_a_at 1449055_x_at | 1532 | 0.837 | 0.3133 | Yes | ||
| 26 | PPARGC1A | 1434099_at 1434100_x_at 1437751_at 1456395_at 1460336_at | 1679 | 0.779 | 0.3141 | Yes | ||
| 27 | CNTFR | 1419429_at 1431663_a_at | 1689 | 0.776 | 0.3212 | Yes | ||
| 28 | MRPS28 | 1452585_at | 1697 | 0.771 | 0.3284 | Yes | ||
| 29 | PDLIM7 | 1417959_at 1428319_at 1443785_x_at | 1961 | 0.680 | 0.3229 | Yes | ||
| 30 | RQCD1 | 1450887_at | 2076 | 0.646 | 0.3239 | Yes | ||
| 31 | PAPD4 | 1419183_at 1433568_at | 2137 | 0.631 | 0.3272 | Yes | ||
| 32 | PALM | 1422034_a_at 1423967_at 1439927_at 1457217_at | 2143 | 0.629 | 0.3331 | Yes | ||
| 33 | WBSCR16 | 1416622_at | 2177 | 0.620 | 0.3376 | Yes | ||
| 34 | NR2F1 | 1418157_at 1422412_x_at | 2215 | 0.608 | 0.3418 | Yes | ||
| 35 | NR2F6 | 1449846_at 1460647_a_at 1460648_at | 2231 | 0.604 | 0.3469 | Yes | ||
| 36 | NDUFS1 | 1425143_a_at 1453354_at | 2233 | 0.603 | 0.3527 | Yes | ||
| 37 | CKS1B | 1416698_a_at 1439514_at 1448441_at 1456268_at | 2273 | 0.595 | 0.3567 | Yes | ||
| 38 | AHCYL1 | 1425576_at 1426830_a_at 1426831_at | 2295 | 0.590 | 0.3614 | Yes | ||
| 39 | CXCL14 | 1418456_a_at 1418457_at | 2382 | 0.572 | 0.3630 | Yes | ||
| 40 | HSPE1 | 1450668_s_at | 2429 | 0.562 | 0.3664 | Yes | ||
| 41 | PDZK1 | 1431701_a_at | 2587 | 0.528 | 0.3643 | Yes | ||
| 42 | HMGCS2 | 1423858_a_at 1431833_a_at | 2655 | 0.513 | 0.3662 | Yes | ||
| 43 | RNF139 | 1429425_at 1429426_at | 2714 | 0.500 | 0.3683 | Yes | ||
| 44 | TRAF4 | 1416571_at 1460642_at | 2729 | 0.498 | 0.3725 | Yes | ||
| 45 | REST | 1425564_at 1425565_at 1428227_at | 2741 | 0.495 | 0.3768 | Yes | ||
| 46 | KIRREL3 | 1431402_at 1441357_at 1452728_at | 2815 | 0.478 | 0.3781 | Yes | ||
| 47 | SHC1 | 1422853_at 1422854_at | 3459 | 0.392 | 0.3523 | No | ||
| 48 | SAP18 | 1419443_at 1419444_at 1419445_s_at 1434646_s_at 1449480_at | 3497 | 0.388 | 0.3544 | No | ||
| 49 | STMN2 | 1423280_at 1423281_at | 3532 | 0.384 | 0.3565 | No | ||
| 50 | HOXD3 | 1421537_at 1437664_at | 3818 | 0.352 | 0.3469 | No | ||
| 51 | SCNM1 | 1428364_at | 4016 | 0.335 | 0.3411 | No | ||
| 52 | AGPS | 1443797_at 1454918_at | 4133 | 0.326 | 0.3389 | No | ||
| 53 | HOXA3 | 1427433_s_at 1441070_at 1452421_at | 4175 | 0.323 | 0.3401 | No | ||
| 54 | ATP5O | 1416278_a_at | 4431 | 0.305 | 0.3314 | No | ||
| 55 | FOXA1 | 1418496_at | 4549 | 0.297 | 0.3289 | No | ||
| 56 | FABP1 | 1417556_at 1448764_a_at | 4592 | 0.294 | 0.3298 | No | ||
| 57 | ZIC4 | 1421539_at 1456417_at | 5095 | 0.264 | 0.3093 | No | ||
| 58 | OBSCN | 1443632_at | 5163 | 0.260 | 0.3087 | No | ||
| 59 | CRABP2 | 1451191_at | 5341 | 0.252 | 0.3030 | No | ||
| 60 | PTHR1 | 1417092_at | 5457 | 0.246 | 0.3001 | No | ||
| 61 | SS18 | 1419360_a_at 1419361_at 1436257_at 1458684_at | 5517 | 0.242 | 0.2998 | No | ||
| 62 | SLC26A3 | 1421445_at 1427547_a_at 1429467_s_at 1453151_at | 5565 | 0.240 | 0.2999 | No | ||
| 63 | CLCNKA | 1450182_at | 5823 | 0.228 | 0.2903 | No | ||
| 64 | THPO | 1449569_at | 5860 | 0.227 | 0.2909 | No | ||
| 65 | CYP26A1 | 1419430_at | 6006 | 0.220 | 0.2863 | No | ||
| 66 | EFEMP1 | 1427183_at | 6130 | 0.215 | 0.2828 | No | ||
| 67 | POLR2H | 1424473_at | 6184 | 0.213 | 0.2824 | No | ||
| 68 | PRODH2 | 1432099_a_at | 6789 | 0.190 | 0.2565 | No | ||
| 69 | PRDM16 | 1429308_at 1429309_at 1440745_at 1440870_at 1443251_at | 6970 | 0.183 | 0.2500 | No | ||
| 70 | PNOC | 1425892_a_at | 7017 | 0.182 | 0.2496 | No | ||
| 71 | CLSPN | 1441984_at 1456280_at | 7044 | 0.181 | 0.2502 | No | ||
| 72 | PAPLN | 1420521_at | 7279 | 0.173 | 0.2411 | No | ||
| 73 | NR2C1 | 1418605_at 1439490_at 1446194_at 1449157_at 1458338_x_at | 7726 | 0.158 | 0.2221 | No | ||
| 74 | WTAP | 1438471_at 1452401_at 1454805_at | 7778 | 0.156 | 0.2213 | No | ||
| 75 | ATP2B1 | 1428936_at 1428937_at 1452969_at 1458346_x_at | 7928 | 0.151 | 0.2159 | No | ||
| 76 | SLC39A5 | 1429523_a_at | 8209 | 0.142 | 0.2044 | No | ||
| 77 | PAX6 | 1419271_at 1425960_s_at 1437816_at 1452526_a_at 1456342_at | 8486 | 0.134 | 0.1930 | No | ||
| 78 | MICAL2 | 1438726_at 1455685_at | 8541 | 0.132 | 0.1918 | No | ||
| 79 | RRBP1 | 1426123_a_at 1436660_at 1449221_a_at 1452767_at | 8743 | 0.125 | 0.1838 | No | ||
| 80 | CLDN15 | 1418920_at | 8950 | 0.119 | 0.1755 | No | ||
| 81 | PGF | 1418471_at | 9567 | 0.100 | 0.1481 | No | ||
| 82 | NRAP | 1421253_at | 9603 | 0.099 | 0.1475 | No | ||
| 83 | IRX6 | 1427383_at | 9631 | 0.099 | 0.1472 | No | ||
| 84 | DCTN2 | 1424461_at | 9715 | 0.096 | 0.1443 | No | ||
| 85 | SMAD3 | 1450471_at 1450472_s_at 1454960_at | 9779 | 0.094 | 0.1423 | No | ||
| 86 | EEF1B2 | 1448252_a_at | 10092 | 0.085 | 0.1288 | No | ||
| 87 | FOXA3 | 1431900_a_at | 10636 | 0.070 | 0.1045 | No | ||
| 88 | ALPI | 1431077_at | 10736 | 0.067 | 0.1006 | No | ||
| 89 | PIPOX | 1449374_at | 10779 | 0.065 | 0.0993 | No | ||
| 90 | RBBP6 | 1425114_at 1425115_at 1425421_at 1426487_a_at 1453611_at | 10931 | 0.061 | 0.0930 | No | ||
| 91 | EIF4EBP2 | 1417084_at 1436158_at 1437733_at 1455575_at 1456613_at | 10978 | 0.060 | 0.0915 | No | ||
| 92 | GDNF | 1419080_at | 11000 | 0.059 | 0.0911 | No | ||
| 93 | SLC23A3 | 1460042_at | 11440 | 0.047 | 0.0713 | No | ||
| 94 | EFNB3 | 1423085_at | 11597 | 0.042 | 0.0646 | No | ||
| 95 | SALL1 | 1450489_at | 11673 | 0.040 | 0.0615 | No | ||
| 96 | ZBTB12 | 1436382_at | 11685 | 0.040 | 0.0614 | No | ||
| 97 | TTYH1 | 1422694_at 1422696_at 1426617_a_at | 11707 | 0.039 | 0.0608 | No | ||
| 98 | PRKCSH | 1416339_a_at | 11769 | 0.037 | 0.0584 | No | ||
| 99 | FXYD1 | 1421374_a_at | 12070 | 0.028 | 0.0449 | No | ||
| 100 | F12 | 1420496_at | 12229 | 0.023 | 0.0378 | No | ||
| 101 | MAP2K3 | 1425456_a_at 1451714_a_at | 12398 | 0.018 | 0.0303 | No | ||
| 102 | ADAMTS19 | 1442459_at 1446513_at | 12514 | 0.015 | 0.0251 | No | ||
| 103 | SYTL2 | 1421594_a_at 1456593_at | 12568 | 0.013 | 0.0228 | No | ||
| 104 | PRDM1 | 1420425_at 1444997_at | 12691 | 0.010 | 0.0173 | No | ||
| 105 | LMO3 | 1455754_at | 12797 | 0.007 | 0.0126 | No | ||
| 106 | GLG1 | 1417087_at 1448579_at 1448580_at 1460554_s_at | 12823 | 0.006 | 0.0115 | No | ||
| 107 | PPP1R14D | 1417990_at | 13099 | -0.004 | -0.0011 | No | ||
| 108 | RBMS1 | 1418703_at 1434005_at 1441437_at 1458578_at 1459968_at | 13184 | -0.007 | -0.0049 | No | ||
| 109 | FGD2 | 1419515_at | 13335 | -0.012 | -0.0117 | No | ||
| 110 | IBRDC2 | 1425282_at 1439153_at 1443252_at 1456834_at | 13352 | -0.012 | -0.0123 | No | ||
| 111 | TPM2 | 1419738_a_at 1419739_at 1425028_a_at 1449577_x_at | 13437 | -0.016 | -0.0160 | No | ||
| 112 | LCAT | 1417043_at | 14041 | -0.037 | -0.0434 | No | ||
| 113 | SLC16A9 | 1429726_at 1429727_at 1454104_a_at | 14347 | -0.049 | -0.0569 | No | ||
| 114 | GOLGA4 | 1417674_s_at 1448803_at 1460213_at | 14493 | -0.055 | -0.0631 | No | ||
| 115 | ERBB2 | 1424919_at | 14627 | -0.060 | -0.0686 | No | ||
| 116 | INSR | 1421380_at 1450225_at | 14980 | -0.075 | -0.0841 | No | ||
| 117 | PRRX2 | 1432331_a_at | 15086 | -0.080 | -0.0881 | No | ||
| 118 | LRP1 | 1442849_at 1448655_at | 15680 | -0.109 | -0.1143 | No | ||
| 119 | POU5F1 | 1417945_at | 15758 | -0.114 | -0.1167 | No | ||
| 120 | GPR124 | 1418379_s_at | 15913 | -0.123 | -0.1226 | No | ||
| 121 | ZSWIM3 | 1428313_at 1444571_at 1457785_at | 15947 | -0.125 | -0.1229 | No | ||
| 122 | ASXL1 | 1434561_at 1435077_at 1458380_at | 16231 | -0.144 | -0.1346 | No | ||
| 123 | PRKCD | 1422847_a_at 1442256_at | 16427 | -0.157 | -0.1420 | No | ||
| 124 | BIN3 | 1417691_at | 16443 | -0.158 | -0.1412 | No | ||
| 125 | HOXC13 | 1425874_at | 16495 | -0.162 | -0.1419 | No | ||
| 126 | TNRC4 | 1452851_at | 16569 | -0.168 | -0.1437 | No | ||
| 127 | CSF2 | 1427429_at | 16572 | -0.169 | -0.1421 | No | ||
| 128 | HSD17B8 | 1454987_a_at | 16613 | -0.171 | -0.1423 | No | ||
| 129 | KLK6 | 1448982_at | 16686 | -0.179 | -0.1439 | No | ||
| 130 | OVOL1 | 1419051_at 1419052_at | 16842 | -0.191 | -0.1491 | No | ||
| 131 | PRKACA | 1447720_x_at 1450519_a_at | 16859 | -0.193 | -0.1480 | No | ||
| 132 | PAX7 | 1444596_at 1452510_at | 16924 | -0.199 | -0.1490 | No | ||
| 133 | RREB1 | 1428657_at 1434741_at 1438216_at 1442047_at 1452862_at | 17073 | -0.214 | -0.1538 | No | ||
| 134 | CLCN2 | 1449248_at | 17197 | -0.227 | -0.1572 | No | ||
| 135 | SYMPK | 1428673_at | 17332 | -0.242 | -0.1610 | No | ||
| 136 | MAGMAS | 1416765_s_at | 17393 | -0.249 | -0.1614 | No | ||
| 137 | MRPL11 | 1417918_at 1437131_x_at 1437839_x_at 1438080_at | 17448 | -0.254 | -0.1614 | No | ||
| 138 | GBL | 1415842_at 1415843_at | 17464 | -0.256 | -0.1596 | No | ||
| 139 | KCNE1L | 1416640_at | 17546 | -0.267 | -0.1607 | No | ||
| 140 | SLC26A6 | 1416275_at | 17615 | -0.275 | -0.1612 | No | ||
| 141 | SFRS4 | 1448778_at | 17648 | -0.278 | -0.1600 | No | ||
| 142 | PKP3 | 1418831_at 1418832_at | 17754 | -0.291 | -0.1620 | No | ||
| 143 | USP37 | 1434242_at 1438212_at | 17828 | -0.299 | -0.1624 | No | ||
| 144 | NCDN | 1416852_a_at 1416853_at | 17884 | -0.307 | -0.1620 | No | ||
| 145 | NR2F2 | 1416158_at 1416159_at 1416160_at 1431237_at 1444229_at | 18287 | -0.365 | -0.1769 | No | ||
| 146 | PFN1 | 1449018_at | 18536 | -0.403 | -0.1844 | No | ||
| 147 | PAK4 | 1428548_at | 18553 | -0.405 | -0.1812 | No | ||
| 148 | PRKAG1 | 1417690_at 1433533_x_at 1457803_at | 18572 | -0.408 | -0.1781 | No | ||
| 149 | YBX2 | 1420762_a_at | 18577 | -0.408 | -0.1743 | No | ||
| 150 | APEX2 | 1424322_at 1425954_a_at | 18637 | -0.421 | -0.1730 | No | ||
| 151 | COMMD3 | 1433539_at 1454642_a_at | 18792 | -0.454 | -0.1757 | No | ||
| 152 | ESRRA | 1442864_at 1460652_at | 19001 | -0.499 | -0.1804 | No | ||
| 153 | AGPAT1 | 1421024_at 1421025_at | 19049 | -0.510 | -0.1776 | No | ||
| 154 | DCTN1 | 1422521_at 1435414_s_at | 19102 | -0.521 | -0.1749 | No | ||
| 155 | GABARAPL2 | 1423187_at 1441376_at 1444956_at 1459477_at | 19131 | -0.528 | -0.1711 | No | ||
| 156 | ARHGEF7 | 1424482_at 1449066_a_at | 19372 | -0.590 | -0.1764 | No | ||
| 157 | AP1B1 | 1416279_at | 19482 | -0.615 | -0.1755 | No | ||
| 158 | ACTA1 | 1427735_a_at | 19508 | -0.620 | -0.1706 | No | ||
| 159 | SCOTIN | 1423986_a_at 1437503_a_at | 19580 | -0.640 | -0.1677 | No | ||
| 160 | HDAC11 | 1451229_at 1454803_a_at | 19584 | -0.641 | -0.1616 | No | ||
| 161 | MAP3K11 | 1450669_at | 19613 | -0.647 | -0.1567 | No | ||
| 162 | MYH10 | 1441057_at 1452740_at | 19960 | -0.765 | -0.1651 | No | ||
| 163 | PCTK1 | 1415956_a_at 1438314_at 1443296_at 1460169_a_at | 20213 | -0.853 | -0.1685 | No | ||
| 164 | RNF5 | 1422888_at | 20268 | -0.876 | -0.1625 | No | ||
| 165 | ZYX | 1417240_at 1438552_x_at | 20494 | -0.981 | -0.1633 | No | ||
| 166 | TTR | 1451580_a_at 1454608_x_at 1455913_x_at 1459737_s_at | 20505 | -0.988 | -0.1542 | No | ||
| 167 | USP52 | 1426700_a_at | 20584 | -1.037 | -0.1477 | No | ||
| 168 | ARF6 | 1418822_a_at 1418823_at 1418824_at | 20682 | -1.104 | -0.1415 | No | ||
| 169 | ETFDH | 1451084_at 1458152_at | 20855 | -1.221 | -0.1376 | No | ||
| 170 | RFX1 | 1422109_at 1436059_at | 20879 | -1.243 | -0.1266 | No | ||
| 171 | DMD | 1417307_at 1430320_at 1446156_at 1448665_at 1457022_at | 20881 | -1.246 | -0.1146 | No | ||
| 172 | LMAN2L | 1435475_at 1441295_at | 21015 | -1.369 | -0.1074 | No | ||
| 173 | NR4A1 | 1416505_at | 21368 | -1.829 | -0.1059 | No | ||
| 174 | TOM1L2 | 1425659_at 1434385_at 1437915_at 1441586_at 1451795_at | 21556 | -2.213 | -0.0930 | No | ||
| 175 | HIBADH | 1423780_at 1435967_s_at 1442848_at 1446297_at | 21610 | -2.375 | -0.0725 | No | ||
| 176 | GBF1 | 1415709_s_at 1415757_at 1432677_at 1438207_at | 21611 | -2.381 | -0.0494 | No | ||
| 177 | HIP2 | 1417186_at 1417187_at 1417188_s_at 1441012_at 1442671_at 1443148_at 1444212_at | 21647 | -2.473 | -0.0271 | No | ||
| 178 | PSMF1 | 1424369_at 1424370_s_at 1424371_at 1451337_at | 21890 | -4.152 | 0.0020 | No |