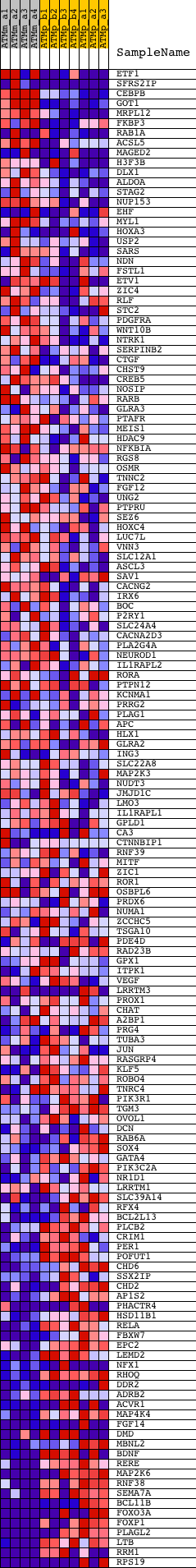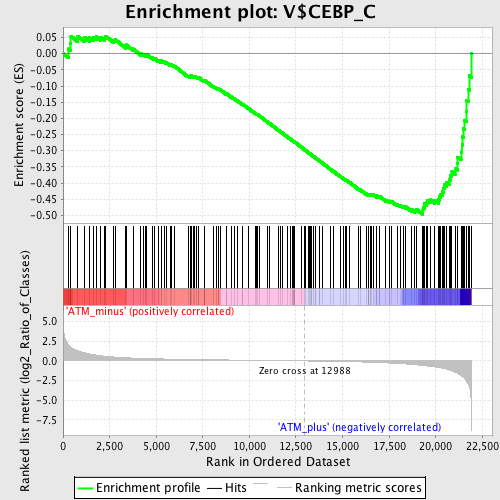
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | V$CEBP_C |
| Enrichment Score (ES) | -0.4971856 |
| Normalized Enrichment Score (NES) | -1.5962384 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.07257804 |
| FWER p-Value | 0.544 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ETF1 | 1420023_at 1420024_s_at 1424013_at 1440629_at 1451208_at | 277 | 2.102 | 0.0146 | No | ||
| 2 | SFRS2IP | 1434691_at 1443085_at 1452884_at 1452885_at 1460445_at | 385 | 1.796 | 0.0330 | No | ||
| 3 | CEBPB | 1418901_at 1427844_a_at | 421 | 1.731 | 0.0539 | No | ||
| 4 | GOT1 | 1450970_at | 795 | 1.271 | 0.0533 | No | ||
| 5 | MRPL12 | 1452048_at | 1147 | 1.030 | 0.0505 | No | ||
| 6 | FKBP3 | 1416858_a_at 1416859_at 1444164_at | 1417 | 0.881 | 0.0496 | No | ||
| 7 | RAB1A | 1416082_at 1448210_at 1457537_at | 1640 | 0.795 | 0.0497 | No | ||
| 8 | ACSL5 | 1428082_at | 1772 | 0.743 | 0.0534 | No | ||
| 9 | MAGED2 | 1426306_a_at 1459873_x_at | 2031 | 0.659 | 0.0501 | No | ||
| 10 | H3F3B | 1420376_a_at 1430357_at 1455725_a_at | 2247 | 0.601 | 0.0480 | No | ||
| 11 | DLX1 | 1449470_at | 2279 | 0.594 | 0.0543 | No | ||
| 12 | ALDOA | 1416921_x_at 1433604_x_at 1434799_x_at | 2719 | 0.500 | 0.0406 | No | ||
| 13 | STAG2 | 1421849_at 1450396_at | 2813 | 0.478 | 0.0426 | No | ||
| 14 | NUP153 | 1441689_at 1441733_s_at 1452176_at | 3365 | 0.403 | 0.0225 | No | ||
| 15 | EHF | 1419474_a_at 1419475_a_at 1447760_x_at 1451375_at | 3380 | 0.401 | 0.0271 | No | ||
| 16 | MYL1 | 1452651_a_at | 3756 | 0.359 | 0.0145 | No | ||
| 17 | HOXA3 | 1427433_s_at 1441070_at 1452421_at | 4175 | 0.323 | -0.0005 | No | ||
| 18 | USP2 | 1417168_a_at 1417169_at | 4307 | 0.313 | -0.0024 | No | ||
| 19 | SARS | 1426257_a_at 1452000_s_at | 4446 | 0.304 | -0.0048 | No | ||
| 20 | NDN | 1415923_at 1435382_at 1435383_x_at 1437853_x_at 1438939_x_at 1438978_x_at 1455792_x_at 1456575_at | 4481 | 0.302 | -0.0024 | No | ||
| 21 | FSTL1 | 1416221_at 1448259_at | 4787 | 0.281 | -0.0128 | No | ||
| 22 | ETV1 | 1422607_at 1445314_at 1450684_at | 4891 | 0.277 | -0.0139 | No | ||
| 23 | ZIC4 | 1421539_at 1456417_at | 5095 | 0.264 | -0.0198 | No | ||
| 24 | RLF | 1427171_at 1439555_at 1440020_at 1446057_at 1447164_at | 5275 | 0.255 | -0.0247 | No | ||
| 25 | STC2 | 1419503_at 1445186_at 1449484_at | 5284 | 0.255 | -0.0218 | No | ||
| 26 | PDGFRA | 1421916_at 1421917_at 1438946_at | 5418 | 0.248 | -0.0247 | No | ||
| 27 | WNT10B | 1426091_a_at | 5526 | 0.242 | -0.0264 | No | ||
| 28 | NTRK1 | 1443990_at | 5774 | 0.231 | -0.0348 | No | ||
| 29 | SERPINB2 | 1419082_at | 5825 | 0.228 | -0.0341 | No | ||
| 30 | CTGF | 1416953_at | 5978 | 0.221 | -0.0382 | No | ||
| 31 | CHST9 | 1431897_at | 6708 | 0.193 | -0.0692 | No | ||
| 32 | CREB5 | 1440089_at 1442576_at 1457222_at | 6831 | 0.188 | -0.0723 | No | ||
| 33 | NOSIP | 1418965_at 1431428_a_at | 6846 | 0.188 | -0.0705 | No | ||
| 34 | RARB | 1454906_at | 6847 | 0.188 | -0.0681 | No | ||
| 35 | GLRA3 | 1450239_at | 6906 | 0.185 | -0.0683 | No | ||
| 36 | PTAFR | 1427871_at 1427872_at | 7006 | 0.182 | -0.0705 | No | ||
| 37 | MEIS1 | 1440431_at 1443260_at 1445773_at 1450992_a_at 1459423_at 1459512_at | 7051 | 0.181 | -0.0702 | No | ||
| 38 | HDAC9 | 1421306_a_at 1434572_at | 7148 | 0.177 | -0.0723 | No | ||
| 39 | NFKBIA | 1420088_at 1420089_at 1438157_s_at 1448306_at 1449731_s_at | 7278 | 0.173 | -0.0760 | No | ||
| 40 | RGS8 | 1453060_at | 7290 | 0.173 | -0.0742 | No | ||
| 41 | OSMR | 1418674_at 1418675_at 1459217_at | 7581 | 0.163 | -0.0854 | No | ||
| 42 | TNNC2 | 1417464_at | 7583 | 0.163 | -0.0834 | No | ||
| 43 | FGF12 | 1440270_at 1451693_a_at 1460137_at | 7612 | 0.162 | -0.0826 | No | ||
| 44 | UNG2 | 1455114_at | 8060 | 0.146 | -0.1012 | No | ||
| 45 | PTPRU | 1416674_at 1446638_at | 8210 | 0.142 | -0.1062 | No | ||
| 46 | SEZ6 | 1420885_a_at 1427674_a_at 1459972_x_at | 8317 | 0.139 | -0.1092 | No | ||
| 47 | HOXC4 | 1422870_at | 8342 | 0.138 | -0.1085 | No | ||
| 48 | LUC7L | 1428255_at 1431181_a_at 1431664_at 1452708_a_at | 8436 | 0.135 | -0.1111 | No | ||
| 49 | VNN3 | 1420723_at | 8781 | 0.124 | -0.1252 | No | ||
| 50 | SLC12A1 | 1421390_at 1424260_at 1443126_at | 8790 | 0.124 | -0.1240 | No | ||
| 51 | ASCL3 | 1420780_at | 9020 | 0.117 | -0.1330 | No | ||
| 52 | SAV1 | 1416075_at 1448204_at | 9190 | 0.112 | -0.1393 | No | ||
| 53 | CACNG2 | 1420596_at 1440210_at 1453363_at | 9371 | 0.107 | -0.1462 | No | ||
| 54 | IRX6 | 1427383_at | 9631 | 0.099 | -0.1568 | No | ||
| 55 | BOC | 1426869_at 1427516_a_at 1427517_at | 9643 | 0.098 | -0.1560 | No | ||
| 56 | P2RY1 | 1421456_at | 9943 | 0.090 | -0.1686 | No | ||
| 57 | SLC24A4 | 1435206_at | 10310 | 0.079 | -0.1844 | No | ||
| 58 | CACNA2D3 | 1419225_at | 10401 | 0.077 | -0.1875 | No | ||
| 59 | PLA2G4A | 1448558_a_at | 10435 | 0.076 | -0.1880 | No | ||
| 60 | NEUROD1 | 1426412_at 1426413_at | 10562 | 0.072 | -0.1929 | No | ||
| 61 | IL1RAPL2 | 1420462_at 1427645_a_at 1441168_at 1446520_at 1446875_at | 10977 | 0.060 | -0.2111 | No | ||
| 62 | RORA | 1420583_a_at 1424034_at 1424035_at 1436325_at 1436326_at 1441085_at 1443511_at 1443647_at 1446360_at 1455165_at 1457177_at 1458129_at | 11080 | 0.057 | -0.2151 | No | ||
| 63 | PTPN12 | 1422045_a_at 1439705_at 1450478_a_at 1450479_x_at 1455105_at 1459459_at | 11557 | 0.044 | -0.2363 | No | ||
| 64 | KCNMA1 | 1424848_at 1425987_a_at 1440728_at 1444973_at 1445774_at 1457018_at | 11677 | 0.040 | -0.2413 | No | ||
| 65 | PRRG2 | 1424329_a_at 1432763_at | 11766 | 0.038 | -0.2448 | No | ||
| 66 | PLAG1 | 1421745_at 1443943_at | 12073 | 0.028 | -0.2585 | No | ||
| 67 | APC | 1420956_at 1420957_at 1435543_at 1450056_at | 12212 | 0.024 | -0.2646 | No | ||
| 68 | HLX1 | 1418939_at | 12295 | 0.021 | -0.2680 | No | ||
| 69 | GLRA2 | 1434098_at 1443351_at | 12310 | 0.021 | -0.2684 | No | ||
| 70 | ING3 | 1422805_a_at 1422806_x_at 1450760_a_at 1460082_at | 12372 | 0.019 | -0.2710 | No | ||
| 71 | SLC22A8 | 1416966_at 1435418_at | 12394 | 0.018 | -0.2717 | No | ||
| 72 | MAP2K3 | 1425456_a_at 1451714_a_at | 12398 | 0.018 | -0.2716 | No | ||
| 73 | NUDT3 | 1451575_a_at | 12449 | 0.017 | -0.2737 | No | ||
| 74 | JMJD1C | 1426900_at 1439998_at 1441592_at 1448049_at 1458521_at | 12451 | 0.017 | -0.2735 | No | ||
| 75 | LMO3 | 1455754_at | 12797 | 0.007 | -0.2893 | No | ||
| 76 | IL1RAPL1 | 1445821_at 1458964_at | 12940 | 0.002 | -0.2958 | No | ||
| 77 | GPLD1 | 1418050_at | 13034 | -0.002 | -0.3000 | No | ||
| 78 | CA3 | 1430584_s_at 1449434_at 1453588_at 1460256_at | 13178 | -0.006 | -0.3065 | No | ||
| 79 | CTNNBIP1 | 1417567_at 1431694_a_at | 13207 | -0.008 | -0.3077 | No | ||
| 80 | RNF39 | 1440912_at 1441793_at | 13258 | -0.009 | -0.3098 | No | ||
| 81 | MITF | 1422025_at 1455214_at | 13334 | -0.012 | -0.3131 | No | ||
| 82 | ZIC1 | 1423477_at 1439627_at | 13344 | -0.012 | -0.3134 | No | ||
| 83 | ROR1 | 1429312_s_at 1429313_at 1442067_at 1451014_at 1457413_at | 13467 | -0.016 | -0.3188 | No | ||
| 84 | OSBPL6 | 1438717_a_at 1449627_at 1451831_at 1456495_s_at 1457881_at | 13544 | -0.019 | -0.3220 | No | ||
| 85 | PRDX6 | 1423223_a_at 1442878_at | 13564 | -0.020 | -0.3226 | No | ||
| 86 | NUMA1 | 1423974_at 1423975_s_at 1441090_at 1444576_at 1444577_x_at | 13771 | -0.028 | -0.3317 | No | ||
| 87 | ZCCHC5 | 1437355_at | 13779 | -0.028 | -0.3317 | No | ||
| 88 | TSGA10 | 1436045_at 1447283_at | 13903 | -0.032 | -0.3369 | No | ||
| 89 | PDE4D | 1443289_at 1458212_at 1459289_at 1459311_at | 14361 | -0.049 | -0.3573 | No | ||
| 90 | RAD23B | 1424222_s_at 1450903_at 1455420_at 1456822_at | 14537 | -0.056 | -0.3646 | No | ||
| 91 | GPX1 | 1460671_at | 14878 | -0.070 | -0.3793 | No | ||
| 92 | ITPK1 | 1426733_at | 15064 | -0.078 | -0.3867 | No | ||
| 93 | VEGF | 1420909_at 1451959_a_at | 15178 | -0.085 | -0.3908 | No | ||
| 94 | LRRTM3 | 1434759_at 1434760_at 1434761_at 1440112_at 1444477_at | 15215 | -0.087 | -0.3914 | No | ||
| 95 | PROX1 | 1421336_at 1437894_at 1457432_at | 15380 | -0.095 | -0.3977 | No | ||
| 96 | CHAT | 1440070_at | 15882 | -0.121 | -0.4191 | No | ||
| 97 | A2BP1 | 1418314_a_at 1438217_at 1447735_x_at 1455358_at 1456921_at | 15957 | -0.125 | -0.4209 | No | ||
| 98 | PRG4 | 1449824_at | 16274 | -0.146 | -0.4335 | No | ||
| 99 | TUBA3 | 1438611_at | 16378 | -0.154 | -0.4362 | No | ||
| 100 | JUN | 1417409_at 1448694_at | 16418 | -0.157 | -0.4359 | No | ||
| 101 | RASGRP4 | 1425380_at | 16421 | -0.157 | -0.4340 | No | ||
| 102 | KLF5 | 1430255_at 1451021_a_at 1451739_at | 16500 | -0.163 | -0.4355 | No | ||
| 103 | ROBO4 | 1426206_at | 16525 | -0.165 | -0.4344 | No | ||
| 104 | TNRC4 | 1452851_at | 16569 | -0.168 | -0.4342 | No | ||
| 105 | PIK3R1 | 1425514_at 1425515_at 1438682_at 1444591_at 1451737_at | 16643 | -0.175 | -0.4353 | No | ||
| 106 | TGM3 | 1421355_at 1440150_at | 16826 | -0.190 | -0.4412 | No | ||
| 107 | OVOL1 | 1419051_at 1419052_at | 16842 | -0.191 | -0.4394 | No | ||
| 108 | DCN | 1441506_at 1449368_at | 16968 | -0.203 | -0.4425 | No | ||
| 109 | RAB6A | 1447776_x_at 1448304_a_at 1448305_at | 16999 | -0.207 | -0.4412 | No | ||
| 110 | SOX4 | 1455867_at 1458332_x_at | 17325 | -0.242 | -0.4530 | No | ||
| 111 | GATA4 | 1418863_at 1418864_at 1441364_at | 17510 | -0.261 | -0.4580 | No | ||
| 112 | PIK3C2A | 1421023_at 1425862_a_at 1443655_s_at | 17530 | -0.264 | -0.4555 | No | ||
| 113 | NR1D1 | 1421802_at 1426464_at | 17659 | -0.279 | -0.4577 | No | ||
| 114 | LRRTM1 | 1437746_at 1452624_at 1455883_a_at | 17940 | -0.318 | -0.4664 | No | ||
| 115 | SLC39A14 | 1425649_at 1427035_at 1438490_at 1457770_at | 18103 | -0.339 | -0.4695 | No | ||
| 116 | RFX4 | 1432053_at 1436931_at 1443312_at | 18263 | -0.362 | -0.4721 | No | ||
| 117 | BCL2L13 | 1424406_at 1429539_at | 18383 | -0.376 | -0.4726 | No | ||
| 118 | PLCB2 | 1452481_at | 18698 | -0.433 | -0.4814 | No | ||
| 119 | CRIM1 | 1426951_at 1442548_at 1452253_at 1458497_at 1459632_at | 18893 | -0.475 | -0.4842 | No | ||
| 120 | PER1 | 1449851_at | 18971 | -0.493 | -0.4813 | No | ||
| 121 | POFUT1 | 1443881_at 1450137_at 1456323_at | 19318 | -0.578 | -0.4897 | Yes | ||
| 122 | CHD6 | 1427384_at 1431047_at | 19324 | -0.580 | -0.4824 | Yes | ||
| 123 | SSX2IP | 1417514_at 1420185_at 1448743_at 1449764_x_at | 19328 | -0.580 | -0.4750 | Yes | ||
| 124 | CHD2 | 1444246_at 1445843_at | 19401 | -0.597 | -0.4705 | Yes | ||
| 125 | AP1S2 | 1437998_at 1447903_x_at 1452657_at 1456252_x_at 1460036_at | 19417 | -0.600 | -0.4634 | Yes | ||
| 126 | PHACTR4 | 1432844_at 1434258_s_at 1454380_at 1457369_at | 19532 | -0.628 | -0.4605 | Yes | ||
| 127 | HSD11B1 | 1449038_at | 19562 | -0.635 | -0.4536 | Yes | ||
| 128 | RELA | 1419536_a_at | 19723 | -0.683 | -0.4521 | Yes | ||
| 129 | FBXW7 | 1424986_s_at 1451558_at | 19952 | -0.763 | -0.4526 | Yes | ||
| 130 | EPC2 | 1455317_at | 20170 | -0.838 | -0.4517 | Yes | ||
| 131 | LEMD2 | 1424326_at | 20193 | -0.846 | -0.4417 | Yes | ||
| 132 | NFX1 | 1419752_at 1419753_at 1428248_at 1442397_at | 20287 | -0.885 | -0.4345 | Yes | ||
| 133 | RHOQ | 1427918_a_at 1440925_at | 20363 | -0.914 | -0.4261 | Yes | ||
| 134 | DDR2 | 1422738_at 1455688_at | 20433 | -0.949 | -0.4169 | Yes | ||
| 135 | ADRB2 | 1437302_at | 20462 | -0.963 | -0.4057 | Yes | ||
| 136 | ACVR1 | 1416786_at 1416787_at 1448460_at 1457551_at | 20596 | -1.046 | -0.3982 | Yes | ||
| 137 | MAP4K4 | 1422615_at 1440609_at 1448050_s_at 1459912_at | 20735 | -1.138 | -0.3898 | Yes | ||
| 138 | FGF14 | 1435747_at 1447221_x_at 1447480_at 1449958_a_at 1458830_at 1459073_x_at | 20785 | -1.172 | -0.3768 | Yes | ||
| 139 | DMD | 1417307_at 1430320_at 1446156_at 1448665_at 1457022_at | 20881 | -1.246 | -0.3650 | Yes | ||
| 140 | MBNL2 | 1433754_at 1436858_at 1446475_at 1455827_at 1457237_at 1457477_at | 21048 | -1.411 | -0.3543 | Yes | ||
| 141 | BDNF | 1422168_a_at 1422169_a_at | 21186 | -1.573 | -0.3402 | Yes | ||
| 142 | RERE | 1454670_at | 21197 | -1.582 | -0.3201 | Yes | ||
| 143 | MAP2K6 | 1426850_a_at 1459292_at | 21369 | -1.830 | -0.3042 | Yes | ||
| 144 | RNF38 | 1428101_at 1431993_a_at 1445272_at | 21420 | -1.925 | -0.2815 | Yes | ||
| 145 | SEMA7A | 1422040_at 1459903_at | 21444 | -1.975 | -0.2569 | Yes | ||
| 146 | BCL11B | 1440137_at 1441762_at 1450339_a_at | 21512 | -2.120 | -0.2324 | Yes | ||
| 147 | FOXO3A | 1434831_a_at 1434832_at 1439598_at 1444226_at 1450236_at 1458279_at | 21546 | -2.180 | -0.2056 | Yes | ||
| 148 | FOXP1 | 1421140_a_at 1421141_a_at 1421142_s_at 1435222_at 1438802_at 1443258_at 1446280_at 1447209_at 1455242_at 1456851_at | 21654 | -2.503 | -0.1780 | Yes | ||
| 149 | PLAGL2 | 1417517_at 1417518_at 1417519_at 1442235_at | 21680 | -2.623 | -0.1451 | Yes | ||
| 150 | LTB | 1419135_at | 21767 | -3.034 | -0.1097 | Yes | ||
| 151 | RRM1 | 1415878_at 1440073_at 1448127_at | 21829 | -3.372 | -0.0687 | Yes | ||
| 152 | RPS19 | 1449243_a_at 1460442_at | 21918 | -5.655 | 0.0007 | Yes |