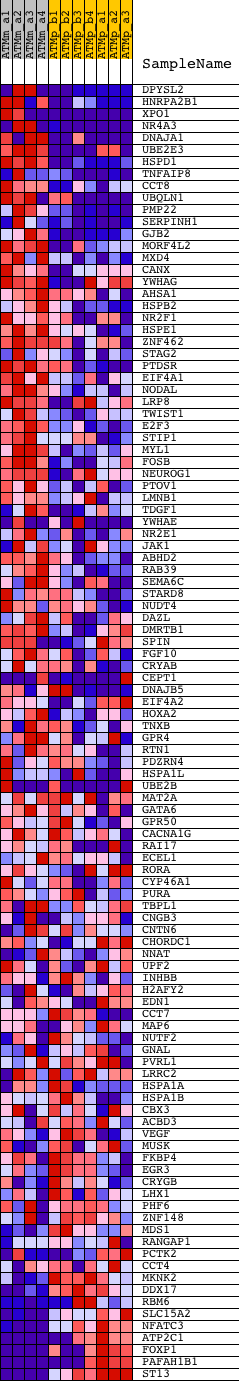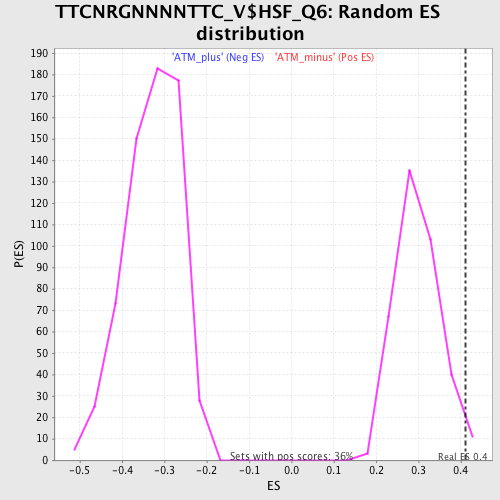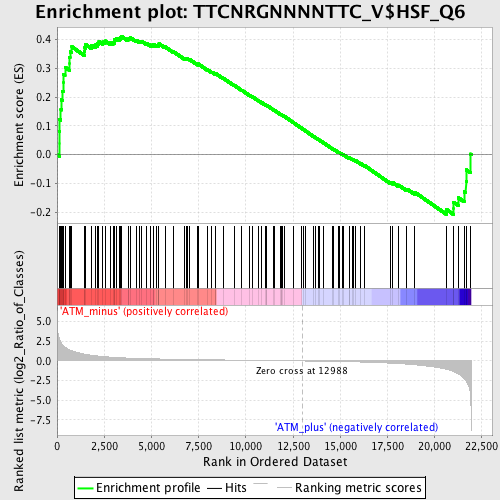
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | TTCNRGNNNNTTC_V$HSF_Q6 |
| Enrichment Score (ES) | 0.41225293 |
| Normalized Enrichment Score (NES) | 1.385063 |
| Nominal p-value | 0.022284122 |
| FDR q-value | 0.32076624 |
| FWER p-Value | 0.987 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | DPYSL2 | 1433770_at 1450502_at 1458188_at 1458632_at | 128 | 2.711 | 0.0374 | Yes | ||
| 2 | HNRPA2B1 | 1420365_a_at 1433829_a_at 1433830_at 1434047_x_at | 129 | 2.707 | 0.0805 | Yes | ||
| 3 | XPO1 | 1418442_at 1418443_at 1448070_at | 141 | 2.630 | 0.1220 | Yes | ||
| 4 | NR4A3 | 1421079_at 1421080_at | 201 | 2.364 | 0.1569 | Yes | ||
| 5 | DNAJA1 | 1416288_at 1437220_x_at 1445729_at 1459835_s_at 1460179_at | 239 | 2.222 | 0.1907 | Yes | ||
| 6 | UBE2E3 | 1448670_at 1448671_at | 292 | 2.033 | 0.2207 | Yes | ||
| 7 | HSPD1 | 1426351_at | 323 | 1.927 | 0.2501 | Yes | ||
| 8 | TNFAIP8 | 1416950_at 1442239_at 1442753_at 1444224_at 1457996_at | 328 | 1.919 | 0.2805 | Yes | ||
| 9 | CCT8 | 1415785_a_at 1436973_at | 419 | 1.732 | 0.3040 | Yes | ||
| 10 | UBQLN1 | 1419385_a_at 1424368_s_at 1450345_at | 650 | 1.414 | 0.3160 | Yes | ||
| 11 | PMP22 | 1417133_at | 653 | 1.408 | 0.3383 | Yes | ||
| 12 | SERPINH1 | 1450843_a_at 1456733_x_at | 685 | 1.379 | 0.3589 | Yes | ||
| 13 | GJB2 | 1423271_at | 782 | 1.286 | 0.3750 | Yes | ||
| 14 | MORF4L2 | 1415778_at 1439413_x_at | 1447 | 0.869 | 0.3584 | Yes | ||
| 15 | MXD4 | 1422574_at 1434378_a_at 1434379_at | 1467 | 0.861 | 0.3713 | Yes | ||
| 16 | CANX | 1415692_s_at 1422845_at 1428935_at | 1491 | 0.854 | 0.3839 | Yes | ||
| 17 | YWHAG | 1420816_at 1420817_at 1450012_x_at 1459693_x_at | 1821 | 0.725 | 0.3804 | Yes | ||
| 18 | AHSA1 | 1424147_at 1444500_at | 2025 | 0.661 | 0.3816 | Yes | ||
| 19 | HSPB2 | 1429888_a_at | 2129 | 0.633 | 0.3870 | Yes | ||
| 20 | NR2F1 | 1418157_at 1422412_x_at | 2215 | 0.608 | 0.3928 | Yes | ||
| 21 | HSPE1 | 1450668_s_at | 2429 | 0.562 | 0.3920 | Yes | ||
| 22 | ZNF462 | 1439106_at 1456789_at | 2541 | 0.537 | 0.3954 | Yes | ||
| 23 | STAG2 | 1421849_at 1450396_at | 2813 | 0.478 | 0.3907 | Yes | ||
| 24 | PTDSR | 1420056_s_at 1420057_at 1454109_a_at | 2961 | 0.455 | 0.3912 | Yes | ||
| 25 | EIF4A1 | 1427058_at 1430980_a_at 1434985_a_at | 3047 | 0.442 | 0.3943 | Yes | ||
| 26 | NODAL | 1422057_at | 3050 | 0.441 | 0.4013 | Yes | ||
| 27 | LRP8 | 1421459_a_at 1440882_at 1442347_at | 3141 | 0.429 | 0.4040 | Yes | ||
| 28 | TWIST1 | 1418733_at | 3313 | 0.408 | 0.4027 | Yes | ||
| 29 | E2F3 | 1427462_at 1434564_at | 3343 | 0.405 | 0.4078 | Yes | ||
| 30 | STIP1 | 1415909_at | 3386 | 0.400 | 0.4123 | Yes | ||
| 31 | MYL1 | 1452651_a_at | 3756 | 0.359 | 0.4011 | No | ||
| 32 | FOSB | 1422134_at | 3800 | 0.354 | 0.4047 | No | ||
| 33 | NEUROG1 | 1438551_at 1450836_at | 3863 | 0.348 | 0.4075 | No | ||
| 34 | PTOV1 | 1416945_at 1438058_s_at 1458357_x_at | 4184 | 0.322 | 0.3979 | No | ||
| 35 | LMNB1 | 1423520_at 1423521_at | 4379 | 0.308 | 0.3940 | No | ||
| 36 | TDGF1 | 1450989_at | 4494 | 0.301 | 0.3935 | No | ||
| 37 | YWHAE | 1426384_a_at 1426385_x_at 1435702_s_at 1438839_a_at 1440841_at 1457173_at | 4726 | 0.285 | 0.3875 | No | ||
| 38 | NR2E1 | 1434921_at 1457289_at | 4951 | 0.272 | 0.3816 | No | ||
| 39 | JAK1 | 1433803_at 1433804_at 1433805_at | 5085 | 0.265 | 0.3797 | No | ||
| 40 | ABHD2 | 1418661_at 1440447_at | 5113 | 0.263 | 0.3827 | No | ||
| 41 | RAB39 | 1437762_at | 5273 | 0.255 | 0.3795 | No | ||
| 42 | SEMA6C | 1423454_a_at 1437652_at 1451898_a_at | 5357 | 0.252 | 0.3797 | No | ||
| 43 | STARD8 | 1427072_at | 5386 | 0.250 | 0.3824 | No | ||
| 44 | NUDT4 | 1418505_at 1449107_at | 5394 | 0.249 | 0.3860 | No | ||
| 45 | DAZL | 1419542_at 1449502_at | 5716 | 0.233 | 0.3750 | No | ||
| 46 | DMRTB1 | 1427252_at | 6140 | 0.215 | 0.3591 | No | ||
| 47 | SPIN | 1415794_a_at 1415795_at 1436809_a_at 1460164_at | 6755 | 0.191 | 0.3340 | No | ||
| 48 | FGF10 | 1420690_at | 6833 | 0.188 | 0.3335 | No | ||
| 49 | CRYAB | 1416455_a_at 1434369_a_at 1435837_at | 6883 | 0.186 | 0.3342 | No | ||
| 50 | CEPT1 | 1416471_at 1443090_at 1446410_at 1459406_at | 6997 | 0.183 | 0.3319 | No | ||
| 51 | DNAJB5 | 1421961_a_at 1421962_at 1450436_s_at | 7441 | 0.167 | 0.3143 | No | ||
| 52 | EIF4A2 | 1450934_at | 7468 | 0.166 | 0.3158 | No | ||
| 53 | HOXA2 | 1419602_at | 7952 | 0.150 | 0.2960 | No | ||
| 54 | TNXB | 1450798_at | 8198 | 0.142 | 0.2871 | No | ||
| 55 | GPR4 | 1457745_at | 8365 | 0.137 | 0.2816 | No | ||
| 56 | RTN1 | 1429761_at | 8375 | 0.137 | 0.2834 | No | ||
| 57 | PDZRN4 | 1456512_at | 8792 | 0.124 | 0.2663 | No | ||
| 58 | HSPA1L | 1419625_at 1440450_at | 9383 | 0.106 | 0.2410 | No | ||
| 59 | UBE2B | 1423106_at 1423107_at 1430177_at 1439477_at 1460132_at | 9787 | 0.094 | 0.2240 | No | ||
| 60 | MAT2A | 1423667_at 1433576_at 1438386_x_at 1438630_x_at 1438976_x_at 1439386_x_at 1456702_x_at | 10198 | 0.082 | 0.2066 | No | ||
| 61 | GATA6 | 1425463_at 1425464_at 1443081_at | 10355 | 0.078 | 0.2006 | No | ||
| 62 | GPR50 | 1450346_at | 10660 | 0.069 | 0.1878 | No | ||
| 63 | CACNA1G | 1423365_at | 10844 | 0.063 | 0.1804 | No | ||
| 64 | RAI17 | 1435740_at 1441823_at 1444505_at 1455984_at 1456816_at 1459305_at | 11010 | 0.059 | 0.1738 | No | ||
| 65 | ECEL1 | 1422586_at | 11075 | 0.057 | 0.1718 | No | ||
| 66 | RORA | 1420583_a_at 1424034_at 1424035_at 1436325_at 1436326_at 1441085_at 1443511_at 1443647_at 1446360_at 1455165_at 1457177_at 1458129_at | 11080 | 0.057 | 0.1725 | No | ||
| 67 | CYP46A1 | 1417709_at | 11482 | 0.046 | 0.1549 | No | ||
| 68 | PURA | 1420628_at 1438219_at 1449934_at 1453783_at 1456898_at | 11506 | 0.045 | 0.1546 | No | ||
| 69 | TBPL1 | 1431415_a_at 1434907_at | 11830 | 0.035 | 0.1403 | No | ||
| 70 | CNGB3 | 1450492_at | 11878 | 0.034 | 0.1387 | No | ||
| 71 | CNTN6 | 1422649_at | 11899 | 0.033 | 0.1383 | No | ||
| 72 | CHORDC1 | 1435574_at 1460645_at | 11927 | 0.032 | 0.1376 | No | ||
| 73 | NNAT | 1423506_a_at | 12023 | 0.029 | 0.1337 | No | ||
| 74 | UPF2 | 1435030_at 1445777_at | 12526 | 0.014 | 0.1110 | No | ||
| 75 | INHBB | 1426858_at 1426859_at | 12943 | 0.002 | 0.0919 | No | ||
| 76 | H2AFY2 | 1426363_x_at 1444008_at 1447241_at | 13062 | -0.003 | 0.0866 | No | ||
| 77 | EDN1 | 1451924_a_at | 13169 | -0.006 | 0.0818 | No | ||
| 78 | CCT7 | 1415816_at 1415817_s_at | 13587 | -0.020 | 0.0630 | No | ||
| 79 | MAP6 | 1422550_a_at 1457316_at | 13699 | -0.025 | 0.0583 | No | ||
| 80 | NUTF2 | 1434367_s_at 1447440_at 1449017_at | 13842 | -0.030 | 0.0523 | No | ||
| 81 | GNAL | 1441654_at 1442021_at 1446359_at 1446491_at | 13915 | -0.033 | 0.0495 | No | ||
| 82 | PVRL1 | 1438111_at 1438421_at 1440131_at 1446488_at 1450819_at | 14092 | -0.040 | 0.0421 | No | ||
| 83 | LRRC2 | 1427388_at 1453628_s_at 1458224_at | 14596 | -0.059 | 0.0200 | No | ||
| 84 | HSPA1A | 1452388_at | 14634 | -0.060 | 0.0193 | No | ||
| 85 | HSPA1B | 1427126_at 1427127_x_at 1452318_a_at | 14890 | -0.070 | 0.0087 | No | ||
| 86 | CBX3 | 1432628_at 1454305_at | 14939 | -0.073 | 0.0077 | No | ||
| 87 | ACBD3 | 1426635_at 1428668_at 1452137_at 1456316_a_at | 15116 | -0.082 | 0.0009 | No | ||
| 88 | VEGF | 1420909_at 1451959_a_at | 15178 | -0.085 | -0.0005 | No | ||
| 89 | MUSK | 1450511_at | 15468 | -0.099 | -0.0122 | No | ||
| 90 | FKBP4 | 1416362_a_at 1416363_at 1458729_at 1459808_at | 15480 | -0.099 | -0.0111 | No | ||
| 91 | EGR3 | 1421486_at 1436329_at | 15625 | -0.106 | -0.0160 | No | ||
| 92 | CRYGB | 1427264_at | 15695 | -0.110 | -0.0174 | No | ||
| 93 | LHX1 | 1421951_at 1450428_at | 15825 | -0.118 | -0.0215 | No | ||
| 94 | PHF6 | 1430415_at 1453761_at 1454625_at | 16083 | -0.133 | -0.0311 | No | ||
| 95 | ZNF148 | 1418381_at 1427730_a_at 1436217_at 1441612_at 1441701_at 1449068_at 1449069_at 1455483_at | 16298 | -0.148 | -0.0386 | No | ||
| 96 | MDS1 | 1422295_at | 17649 | -0.279 | -0.0960 | No | ||
| 97 | RANGAP1 | 1423749_s_at 1444581_at 1451092_a_at | 17786 | -0.295 | -0.0975 | No | ||
| 98 | PCTK2 | 1435143_at 1446130_at 1446272_at | 18078 | -0.335 | -0.1055 | No | ||
| 99 | CCT4 | 1415867_at 1415868_at 1430034_at 1433447_x_at 1438560_x_at 1442569_at 1456082_x_at 1456572_x_at | 18523 | -0.400 | -0.1195 | No | ||
| 100 | MKNK2 | 1418300_a_at 1449029_at | 18946 | -0.487 | -0.1310 | No | ||
| 101 | DDX17 | 1437773_x_at 1439037_at 1452155_a_at | 20612 | -1.055 | -0.1905 | No | ||
| 102 | RBM6 | 1417213_a_at 1440223_at 1452466_a_at 1458912_at | 20997 | -1.350 | -0.1865 | No | ||
| 103 | SLC15A2 | 1417600_at 1424730_a_at 1447808_s_at | 21001 | -1.353 | -0.1651 | No | ||
| 104 | NFATC3 | 1419976_s_at 1452497_a_at | 21248 | -1.658 | -0.1499 | No | ||
| 105 | ATP2C1 | 1431196_at 1434386_at 1437738_at 1439215_at 1442742_at | 21557 | -2.215 | -0.1287 | No | ||
| 106 | FOXP1 | 1421140_a_at 1421141_a_at 1421142_s_at 1435222_at 1438802_at 1443258_at 1446280_at 1447209_at 1455242_at 1456851_at | 21654 | -2.503 | -0.0932 | No | ||
| 107 | PAFAH1B1 | 1417086_at 1427703_at 1439656_at 1441404_at 1444125_at 1448578_at 1456947_at 1460199_a_at | 21667 | -2.566 | -0.0528 | No | ||
| 108 | ST13 | 1416851_at 1442775_at 1460193_at | 21884 | -4.078 | 0.0023 | No |