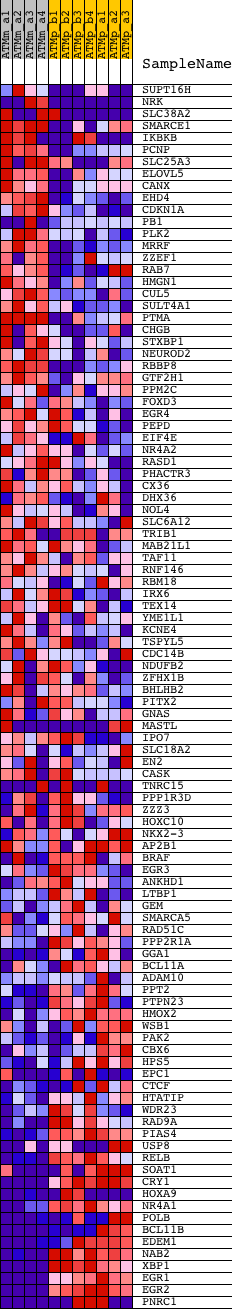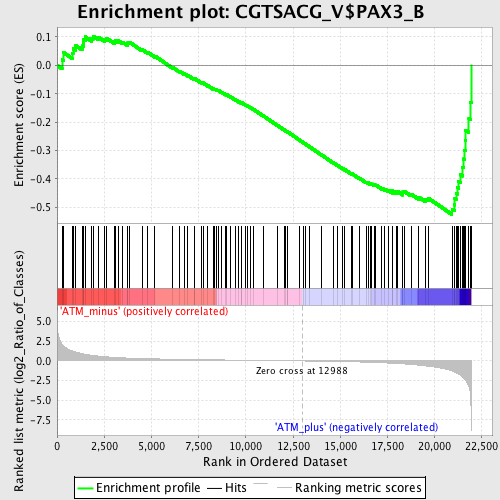
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | CGTSACG_V$PAX3_B |
| Enrichment Score (ES) | -0.5253042 |
| Normalized Enrichment Score (NES) | -1.6188691 |
| Nominal p-value | 0.0015822785 |
| FDR q-value | 0.062158916 |
| FWER p-Value | 0.436 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | SUPT16H | 1419741_at 1449578_at 1456449_at | 263 | 2.147 | 0.0201 | No | ||
| 2 | NRK | 1436399_s_at 1450078_at 1450079_at | 341 | 1.896 | 0.0451 | No | ||
| 3 | SLC38A2 | 1426722_at 1429593_at 1429594_at | 802 | 1.264 | 0.0430 | No | ||
| 4 | SMARCE1 | 1422675_at 1422676_at 1430822_at | 847 | 1.229 | 0.0594 | No | ||
| 5 | IKBKB | 1426207_at 1426333_a_at 1432275_at 1445141_at 1454184_a_at | 991 | 1.128 | 0.0697 | No | ||
| 6 | PCNP | 1428314_at 1452735_at | 1325 | 0.932 | 0.0685 | No | ||
| 7 | SLC25A3 | 1416300_a_at 1440302_at | 1385 | 0.901 | 0.0793 | No | ||
| 8 | ELOVL5 | 1415840_at 1437211_x_at 1459242_at | 1413 | 0.882 | 0.0912 | No | ||
| 9 | CANX | 1415692_s_at 1422845_at 1428935_at | 1491 | 0.854 | 0.1005 | No | ||
| 10 | EHD4 | 1449852_a_at | 1836 | 0.721 | 0.0956 | No | ||
| 11 | CDKN1A | 1421679_a_at 1424638_at | 1903 | 0.701 | 0.1031 | No | ||
| 12 | PB1 | 1426877_a_at 1426878_at 1427266_at 1442603_at | 2184 | 0.618 | 0.0995 | No | ||
| 13 | PLK2 | 1427005_at | 2523 | 0.539 | 0.0921 | No | ||
| 14 | MRRF | 1426364_at | 2625 | 0.519 | 0.0953 | No | ||
| 15 | ZZEF1 | 1434192_at 1438691_at 1446081_at 1453583_at | 3039 | 0.443 | 0.0830 | No | ||
| 16 | RAB7 | 1415734_at 1448159_at | 3067 | 0.438 | 0.0883 | No | ||
| 17 | HMGN1 | 1422495_a_at 1422496_at 1438940_x_at 1455897_x_at | 3233 | 0.419 | 0.0871 | No | ||
| 18 | CUL5 | 1428287_at 1452722_a_at 1456102_a_at | 3472 | 0.390 | 0.0820 | No | ||
| 19 | SULT4A1 | 1421606_a_at 1433714_at | 3738 | 0.360 | 0.0753 | No | ||
| 20 | PTMA | 1423455_at | 3751 | 0.359 | 0.0801 | No | ||
| 21 | CHGB | 1415885_at | 3847 | 0.350 | 0.0810 | No | ||
| 22 | STXBP1 | 1420505_a_at 1420506_a_at | 4505 | 0.300 | 0.0554 | No | ||
| 23 | NEUROD2 | 1444362_at | 4800 | 0.281 | 0.0462 | No | ||
| 24 | RBBP8 | 1427061_at 1427062_at 1446097_at | 5177 | 0.260 | 0.0329 | No | ||
| 25 | GTF2H1 | 1453169_a_at | 6125 | 0.216 | -0.0073 | No | ||
| 26 | PPM2C | 1434228_at 1438201_at | 6480 | 0.201 | -0.0205 | No | ||
| 27 | FOXD3 | 1422210_at | 6732 | 0.192 | -0.0291 | No | ||
| 28 | EGR4 | 1449977_at | 6920 | 0.185 | -0.0349 | No | ||
| 29 | PEPD | 1416712_at | 7266 | 0.173 | -0.0481 | No | ||
| 30 | EIF4E | 1450909_at 1457489_at 1459371_at | 7293 | 0.172 | -0.0467 | No | ||
| 31 | NR4A2 | 1447863_s_at 1450749_a_at 1450750_a_at 1455034_at | 7635 | 0.161 | -0.0599 | No | ||
| 32 | RASD1 | 1423619_at | 7730 | 0.158 | -0.0618 | No | ||
| 33 | PHACTR3 | 1429651_at | 7982 | 0.149 | -0.0711 | No | ||
| 34 | CX36 | 1423019_at 1435297_at | 8280 | 0.140 | -0.0826 | No | ||
| 35 | DHX36 | 1424397_at 1424398_at 1437550_at | 8344 | 0.138 | -0.0834 | No | ||
| 36 | NOL4 | 1430189_at 1441447_at 1456387_at | 8449 | 0.135 | -0.0862 | No | ||
| 37 | SLC6A12 | 1449382_at | 8528 | 0.132 | -0.0878 | No | ||
| 38 | TRIB1 | 1424880_at 1424881_at | 8703 | 0.127 | -0.0938 | No | ||
| 39 | MAB21L1 | 1421369_a_at 1424679_at 1440860_at 1457290_at | 8936 | 0.119 | -0.1027 | No | ||
| 40 | TAF11 | 1446161_at 1451995_at | 8982 | 0.118 | -0.1030 | No | ||
| 41 | RNF146 | 1417733_at 1425001_at 1425713_a_at | 9194 | 0.112 | -0.1109 | No | ||
| 42 | RBM18 | 1420607_at 1420608_at 1458182_at | 9447 | 0.104 | -0.1209 | No | ||
| 43 | IRX6 | 1427383_at | 9631 | 0.099 | -0.1278 | No | ||
| 44 | TEX14 | 1419240_at | 9747 | 0.095 | -0.1317 | No | ||
| 45 | YME1L1 | 1423360_at 1423361_at 1437822_at 1450954_at | 9761 | 0.095 | -0.1308 | No | ||
| 46 | KCNE4 | 1418156_at | 9994 | 0.088 | -0.1401 | No | ||
| 47 | TSPYL5 | 1455807_at | 10086 | 0.085 | -0.1430 | No | ||
| 48 | CDC14B | 1429418_at 1437070_at | 10244 | 0.081 | -0.1490 | No | ||
| 49 | NDUFB2 | 1416834_x_at 1442658_at 1448483_a_at 1453806_at | 10405 | 0.076 | -0.1552 | No | ||
| 50 | ZFHX1B | 1422748_at 1434298_at 1442393_at 1445518_at 1454200_at 1456389_at | 10928 | 0.061 | -0.1782 | No | ||
| 51 | BHLHB2 | 1418025_at | 11657 | 0.041 | -0.2109 | No | ||
| 52 | PITX2 | 1424797_a_at 1450482_a_at | 12020 | 0.029 | -0.2271 | No | ||
| 53 | GNAS | 1421740_at 1427789_s_at 1443007_at 1443375_at 1444767_at 1450186_s_at 1453413_at | 12117 | 0.027 | -0.2311 | No | ||
| 54 | MASTL | 1423524_at 1423525_at 1444725_at 1447136_at | 12174 | 0.025 | -0.2333 | No | ||
| 55 | IPO7 | 1442511_at 1454955_at 1458165_at | 12189 | 0.024 | -0.2335 | No | ||
| 56 | SLC18A2 | 1437079_at | 12837 | 0.005 | -0.2631 | No | ||
| 57 | EN2 | 1418868_at | 13039 | -0.002 | -0.2723 | No | ||
| 58 | CASK | 1422518_at 1422519_at 1427692_a_at 1445152_at | 13139 | -0.005 | -0.2767 | No | ||
| 59 | TNRC15 | 1439837_at 1446275_at 1451856_at 1457472_at | 13383 | -0.014 | -0.2876 | No | ||
| 60 | PPP1R3D | 1452922_at | 13980 | -0.035 | -0.3144 | No | ||
| 61 | ZZZ3 | 1434332_at 1438487_s_at 1441748_at | 14629 | -0.060 | -0.3432 | No | ||
| 62 | HOXC10 | 1439798_at | 14855 | -0.069 | -0.3525 | No | ||
| 63 | NKX2-3 | 1425827_at 1427741_x_at 1452528_a_at | 15133 | -0.082 | -0.3639 | No | ||
| 64 | AP2B1 | 1427077_a_at 1445142_at 1446566_at 1452292_at 1458971_at | 15241 | -0.088 | -0.3675 | No | ||
| 65 | BRAF | 1425693_at 1435434_at 1435480_at 1442749_at 1445786_at 1447940_a_at 1447941_x_at 1456505_at 1458641_at | 15601 | -0.105 | -0.3824 | No | ||
| 66 | EGR3 | 1421486_at 1436329_at | 15625 | -0.106 | -0.3818 | No | ||
| 67 | ANKHD1 | 1419867_a_at 1436597_at 1440754_at 1441597_at 1442641_at 1448008_at 1453023_at 1455759_a_at | 16020 | -0.130 | -0.3979 | No | ||
| 68 | LTBP1 | 1419786_at 1440678_at 1446232_at 1447547_at 1448870_at 1457852_at 1458739_at | 16376 | -0.154 | -0.4119 | No | ||
| 69 | GEM | 1426063_a_at | 16465 | -0.160 | -0.4135 | No | ||
| 70 | SMARCA5 | 1440048_at | 16594 | -0.170 | -0.4168 | No | ||
| 71 | RAD51C | 1423026_at 1438453_at | 16675 | -0.178 | -0.4178 | No | ||
| 72 | PPP2R1A | 1415819_a_at 1438174_x_at 1438383_x_at 1438991_x_at 1455929_x_at | 16787 | -0.186 | -0.4201 | No | ||
| 73 | GGA1 | 1423834_s_at 1459863_x_at | 16878 | -0.194 | -0.4213 | No | ||
| 74 | BCL11A | 1419406_a_at 1426552_a_at 1446293_at 1447334_at 1447335_x_at 1453814_at 1456632_at | 17203 | -0.227 | -0.4328 | No | ||
| 75 | ADAM10 | 1450104_at 1460083_at | 17357 | -0.244 | -0.4361 | No | ||
| 76 | PPT2 | 1418302_at | 17543 | -0.266 | -0.4406 | No | ||
| 77 | PTPN23 | 1427128_at | 17747 | -0.290 | -0.4455 | No | ||
| 78 | HMOX2 | 1416399_a_at | 17769 | -0.293 | -0.4421 | No | ||
| 79 | WSB1 | 1425241_a_at | 17971 | -0.321 | -0.4465 | No | ||
| 80 | PAK2 | 1434250_at 1454887_at | 18043 | -0.329 | -0.4448 | No | ||
| 81 | CBX6 | 1424407_s_at 1429290_at | 18307 | -0.367 | -0.4514 | No | ||
| 82 | HPS5 | 1434677_at | 18312 | -0.368 | -0.4460 | No | ||
| 83 | EPC1 | 1418850_at 1429914_at 1440198_at 1440742_at 1442279_at 1443675_at 1458315_at | 18390 | -0.377 | -0.4439 | No | ||
| 84 | CTCF | 1418330_at 1449042_at | 18744 | -0.444 | -0.4534 | No | ||
| 85 | HTATIP | 1433980_at 1433981_s_at | 19145 | -0.531 | -0.4638 | No | ||
| 86 | WDR23 | 1448418_s_at 1460189_at | 19527 | -0.626 | -0.4718 | No | ||
| 87 | RAD9A | 1418404_at | 19655 | -0.662 | -0.4677 | No | ||
| 88 | PIAS4 | 1418861_at 1455394_at | 20913 | -1.277 | -0.5062 | Yes | ||
| 89 | USP8 | 1422883_at 1443420_at 1446807_at 1447386_at | 21044 | -1.408 | -0.4910 | Yes | ||
| 90 | RELB | 1417856_at | 21067 | -1.442 | -0.4704 | Yes | ||
| 91 | SOAT1 | 1417695_a_at 1417696_at 1417697_at 1426818_at 1448068_at | 21135 | -1.506 | -0.4509 | Yes | ||
| 92 | CRY1 | 1433733_a_at | 21191 | -1.574 | -0.4298 | Yes | ||
| 93 | HOXA9 | 1421579_at 1455626_at | 21257 | -1.668 | -0.4077 | Yes | ||
| 94 | NR4A1 | 1416505_at | 21368 | -1.829 | -0.3853 | Yes | ||
| 95 | POLB | 1425371_at 1434229_a_at | 21467 | -2.021 | -0.3595 | Yes | ||
| 96 | BCL11B | 1440137_at 1441762_at 1450339_a_at | 21512 | -2.120 | -0.3297 | Yes | ||
| 97 | EDEM1 | 1424065_at 1451218_at | 21560 | -2.220 | -0.2986 | Yes | ||
| 98 | NAB2 | 1417930_at | 21625 | -2.418 | -0.2653 | Yes | ||
| 99 | XBP1 | 1420011_s_at 1420012_at 1420886_a_at 1437223_s_at | 21644 | -2.460 | -0.2292 | Yes | ||
| 100 | EGR1 | 1417065_at | 21809 | -3.263 | -0.1878 | Yes | ||
| 101 | EGR2 | 1427682_a_at 1427683_at | 21887 | -4.118 | -0.1295 | Yes | ||
| 102 | PNRC1 | 1433668_at 1438524_x_at 1439513_at | 21934 | -8.778 | -0.0000 | Yes |