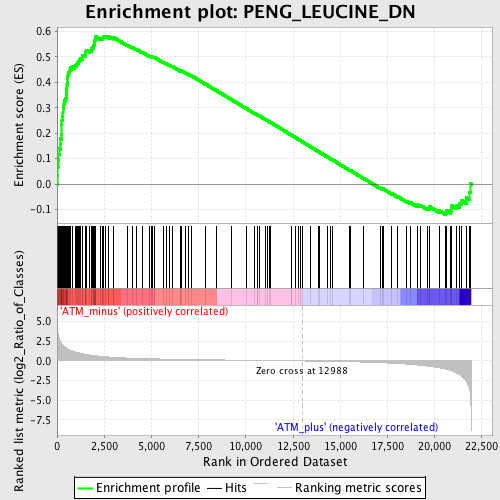
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | PENG_LEUCINE_DN |
| Enrichment Score (ES) | 0.58163524 |
| Normalized Enrichment Score (NES) | 2.0175111 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.004040598 |
| FWER p-Value | 0.0040 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | NCL | 1415771_at 1415772_at 1415773_at 1442404_at 1456528_x_at | 21 | 4.111 | 0.0338 | Yes | ||
| 2 | TCP1 | 1442317_at 1448122_at | 33 | 3.812 | 0.0655 | Yes | ||
| 3 | XPO6 | 1422759_a_at 1440492_at 1443737_at | 49 | 3.490 | 0.0944 | Yes | ||
| 4 | PSMD1 | 1419799_at 1429291_at | 88 | 3.036 | 0.1183 | Yes | ||
| 5 | CYCS | 1422484_at 1445484_at | 133 | 2.683 | 0.1390 | Yes | ||
| 6 | MRPL3 | 1422463_a_at 1422464_at 1431006_at 1440636_at | 179 | 2.479 | 0.1579 | Yes | ||
| 7 | TOMM40 | 1426118_a_at 1459680_at | 184 | 2.454 | 0.1784 | Yes | ||
| 8 | CCNB1 | 1419943_s_at 1419944_at | 221 | 2.298 | 0.1962 | Yes | ||
| 9 | DNAJA1 | 1416288_at 1437220_x_at 1445729_at 1459835_s_at 1460179_at | 239 | 2.222 | 0.2142 | Yes | ||
| 10 | VDAC1 | 1415998_at 1436992_x_at 1437192_x_at 1437452_x_at 1437947_x_at | 240 | 2.219 | 0.2330 | Yes | ||
| 11 | TFRC | 1422966_a_at 1422967_a_at 1452661_at AFFX-TransRecMur/X57349_3_at AFFX-TransRecMur/X57349_5_at AFFX-TransRecMur/X57349_M_at | 250 | 2.192 | 0.2511 | Yes | ||
| 12 | ETF1 | 1420023_at 1420024_s_at 1424013_at 1440629_at 1451208_at | 277 | 2.102 | 0.2677 | Yes | ||
| 13 | XRCC5 | 1451968_at | 310 | 1.958 | 0.2828 | Yes | ||
| 14 | TRIP13 | 1429294_at 1429295_s_at 1446760_at | 312 | 1.957 | 0.2993 | Yes | ||
| 15 | SLC29A1 | 1451782_a_at | 349 | 1.891 | 0.3137 | Yes | ||
| 16 | AMD1 | 1448484_at | 368 | 1.845 | 0.3284 | Yes | ||
| 17 | HPRT1 | 1448736_a_at | 464 | 1.663 | 0.3381 | Yes | ||
| 18 | CTSC | 1416382_at 1437939_s_at 1446834_at | 486 | 1.618 | 0.3509 | Yes | ||
| 19 | NMT1 | 1415683_at 1418082_at 1454941_at | 500 | 1.602 | 0.3638 | Yes | ||
| 20 | DDX21 | 1448270_at 1448271_a_at | 507 | 1.589 | 0.3770 | Yes | ||
| 21 | UBE2N | 1422559_at 1435384_at | 522 | 1.575 | 0.3896 | Yes | ||
| 22 | CCT6A | 1423517_at 1455988_a_at | 552 | 1.536 | 0.4013 | Yes | ||
| 23 | SNRPA1 | 1417351_a_at 1417352_s_at 1417353_x_at | 563 | 1.523 | 0.4137 | Yes | ||
| 24 | MCM4 | 1416214_at 1436708_x_at | 574 | 1.511 | 0.4260 | Yes | ||
| 25 | NOL5A | 1426533_at 1455035_s_at | 591 | 1.483 | 0.4378 | Yes | ||
| 26 | SRM | 1421260_a_at | 634 | 1.429 | 0.4480 | Yes | ||
| 27 | DDX1 | 1415915_at 1440816_x_at | 688 | 1.374 | 0.4572 | Yes | ||
| 28 | TGFB1 | 1420653_at 1445360_at | 817 | 1.249 | 0.4619 | Yes | ||
| 29 | CCT5 | 1417258_at | 947 | 1.148 | 0.4657 | Yes | ||
| 30 | NP | 1416530_a_at | 1017 | 1.110 | 0.4719 | Yes | ||
| 31 | CAD | 1440857_at 1452829_at 1452830_s_at | 1100 | 1.057 | 0.4771 | Yes | ||
| 32 | MRPL12 | 1452048_at | 1147 | 1.030 | 0.4837 | Yes | ||
| 33 | SQLE | 1415993_at | 1203 | 1.003 | 0.4896 | Yes | ||
| 34 | BST2 | 1424921_at | 1260 | 0.968 | 0.4953 | Yes | ||
| 35 | HSPA4 | 1416146_at 1416147_at 1440575_at | 1348 | 0.920 | 0.4991 | Yes | ||
| 36 | EIF4G2 | 1415863_at 1452758_s_at | 1355 | 0.919 | 0.5065 | Yes | ||
| 37 | TK1 | 1416258_at | 1482 | 0.856 | 0.5080 | Yes | ||
| 38 | GLO1 | 1424108_at 1424109_a_at 1436070_at 1451240_a_at | 1495 | 0.853 | 0.5147 | Yes | ||
| 39 | LDHA | 1419737_a_at | 1527 | 0.839 | 0.5203 | Yes | ||
| 40 | TIMM17A | 1426256_at | 1546 | 0.832 | 0.5265 | Yes | ||
| 41 | UNG | 1425753_a_at | 1740 | 0.755 | 0.5241 | Yes | ||
| 42 | ERH | 1430536_a_at | 1811 | 0.729 | 0.5270 | Yes | ||
| 43 | RNF126 | 1424514_at | 1840 | 0.719 | 0.5318 | Yes | ||
| 44 | SSB | 1416421_a_at 1416422_a_at 1416423_x_at 1453928_a_at | 1863 | 0.713 | 0.5369 | Yes | ||
| 45 | VRK1 | 1425006_a_at 1458951_at | 1942 | 0.686 | 0.5391 | Yes | ||
| 46 | CCT2 | 1416037_a_at 1433534_a_at 1433535_x_at | 1946 | 0.684 | 0.5447 | Yes | ||
| 47 | NME2 | 1448808_a_at | 1968 | 0.679 | 0.5495 | Yes | ||
| 48 | RBM14 | 1436979_x_at 1437032_x_at 1443033_at 1450569_a_at 1452003_at 1456566_x_at | 1978 | 0.676 | 0.5548 | Yes | ||
| 49 | EIF4G1 | 1427036_a_at 1427037_at 1438686_at | 1995 | 0.672 | 0.5598 | Yes | ||
| 50 | POLG | 1423272_at 1423273_at 1437564_at | 1998 | 0.671 | 0.5654 | Yes | ||
| 51 | AHSA1 | 1424147_at 1444500_at | 2025 | 0.661 | 0.5698 | Yes | ||
| 52 | NOL1 | 1424019_at | 2045 | 0.654 | 0.5744 | Yes | ||
| 53 | EI24 | 1416555_at | 2050 | 0.653 | 0.5798 | Yes | ||
| 54 | CCNA2 | 1417910_at 1417911_at | 2318 | 0.587 | 0.5725 | Yes | ||
| 55 | MRPL28 | 1416284_at 1437622_x_at 1456313_x_at | 2396 | 0.570 | 0.5738 | Yes | ||
| 56 | HSPE1 | 1450668_s_at | 2429 | 0.562 | 0.5771 | Yes | ||
| 57 | CYC1 | 1416604_at | 2444 | 0.559 | 0.5811 | Yes | ||
| 58 | CSE1L | 1448809_at | 2566 | 0.532 | 0.5801 | Yes | ||
| 59 | SCD | 1415964_at 1415965_at | 2700 | 0.503 | 0.5782 | Yes | ||
| 60 | ALDOA | 1416921_x_at 1433604_x_at 1434799_x_at | 2719 | 0.500 | 0.5816 | Yes | ||
| 61 | PSMA7 | 1423567_a_at 1423568_at | 2969 | 0.453 | 0.5740 | No | ||
| 62 | HSPA5 | 1416064_a_at 1427464_s_at 1447824_x_at | 3009 | 0.447 | 0.5760 | No | ||
| 63 | MRPS12 | 1448799_s_at | 3725 | 0.362 | 0.5463 | No | ||
| 64 | FASN | 1423828_at | 3975 | 0.339 | 0.5377 | No | ||
| 65 | PSMD11 | 1429370_a_at 1432726_at 1456059_at 1456104_at | 4196 | 0.321 | 0.5304 | No | ||
| 66 | PSMD3 | 1448479_at | 4525 | 0.299 | 0.5179 | No | ||
| 67 | SEC61B | 1417083_at | 4905 | 0.276 | 0.5028 | No | ||
| 68 | COPB2 | 1452102_at 1456175_a_at | 5018 | 0.269 | 0.4999 | No | ||
| 69 | EBP | 1416667_at | 5054 | 0.266 | 0.5006 | No | ||
| 70 | NBL1 | 1448428_at | 5145 | 0.262 | 0.4987 | No | ||
| 71 | EIF5 | 1415723_at 1433631_at 1454663_at 1454664_a_at 1456256_at | 5633 | 0.236 | 0.4783 | No | ||
| 72 | BYSL | 1422767_at 1450742_at | 5777 | 0.231 | 0.4737 | No | ||
| 73 | PFKM | 1416780_at | 5961 | 0.222 | 0.4672 | No | ||
| 74 | FDPS | 1423418_at | 6092 | 0.217 | 0.4631 | No | ||
| 75 | DNAJB6 | 1429776_a_at 1429777_at 1434035_at 1435701_at 1442217_at 1448234_at | 6528 | 0.199 | 0.4448 | No | ||
| 76 | SFRS2 | 1415807_s_at 1427504_s_at 1427816_at 1452439_s_at | 6535 | 0.199 | 0.4462 | No | ||
| 77 | ARMET | 1428112_at 1459455_at | 6591 | 0.197 | 0.4454 | No | ||
| 78 | ADRM1 | 1419263_a_at 1438170_x_at | 6819 | 0.189 | 0.4366 | No | ||
| 79 | UBC | 1420494_x_at 1425965_at 1425966_x_at 1432827_x_at 1437666_x_at 1454373_x_at | 6947 | 0.184 | 0.4323 | No | ||
| 80 | PSMB7 | 1416240_at | 7112 | 0.178 | 0.4263 | No | ||
| 81 | NUP98 | 1434220_at 1439154_at 1446397_at 1446988_at | 7848 | 0.154 | 0.3939 | No | ||
| 82 | DHX9 | 1425617_at 1425618_at 1451770_s_at | 8415 | 0.136 | 0.3691 | No | ||
| 83 | PAPOLA | 1424216_a_at 1424217_at 1427544_a_at 1444015_at 1455836_at | 9255 | 0.110 | 0.3315 | No | ||
| 84 | OGFR | 1422511_a_at 1422512_a_at | 10030 | 0.087 | 0.2967 | No | ||
| 85 | EMP3 | 1417104_at | 10454 | 0.075 | 0.2780 | No | ||
| 86 | ACAT2 | 1435630_s_at | 10596 | 0.071 | 0.2721 | No | ||
| 87 | GART | 1416283_at 1424435_a_at 1424436_at 1441911_x_at | 10617 | 0.070 | 0.2718 | No | ||
| 88 | PSMA5 | 1424681_a_at 1434356_a_at | 10708 | 0.068 | 0.2682 | No | ||
| 89 | CALR | 1417606_a_at 1433806_x_at 1456170_x_at | 11055 | 0.058 | 0.2529 | No | ||
| 90 | DHCR24 | 1418129_at 1451895_a_at | 11128 | 0.056 | 0.2500 | No | ||
| 91 | PIK3R3 | 1445097_at 1447068_at 1456482_at | 11259 | 0.053 | 0.2445 | No | ||
| 92 | GSPT1 | 1426736_at 1426737_at 1435000_at 1446550_at 1452168_x_at 1455173_at | 11318 | 0.051 | 0.2423 | No | ||
| 93 | MAP2K3 | 1425456_a_at 1451714_a_at | 12398 | 0.018 | 0.1929 | No | ||
| 94 | EIF2B4 | 1449940_a_at | 12423 | 0.017 | 0.1920 | No | ||
| 95 | GCDH | 1448717_at | 12640 | 0.011 | 0.1822 | No | ||
| 96 | ME2 | 1426572_at 1426573_at 1458172_at | 12782 | 0.007 | 0.1758 | No | ||
| 97 | PPIB | 1437649_x_at 1450911_at | 12913 | 0.003 | 0.1698 | No | ||
| 98 | FKBP2 | 1450694_at | 13010 | -0.001 | 0.1654 | No | ||
| 99 | MLF2 | 1423916_s_at | 13440 | -0.016 | 0.1459 | No | ||
| 100 | NUTF2 | 1434367_s_at 1447440_at 1449017_at | 13842 | -0.030 | 0.1277 | No | ||
| 101 | ILF3 | 1422546_at 1436802_at 1460669_at | 13882 | -0.032 | 0.1262 | No | ||
| 102 | ALG8 | 1455887_at | 14341 | -0.049 | 0.1056 | No | ||
| 103 | DHRS1 | 1415677_at | 14501 | -0.055 | 0.0988 | No | ||
| 104 | NXF1 | 1416791_a_at | 14569 | -0.058 | 0.0962 | No | ||
| 105 | FKBP4 | 1416362_a_at 1416363_at 1458729_at 1459808_at | 15480 | -0.099 | 0.0553 | No | ||
| 106 | PSMD2 | 1415831_at | 15516 | -0.101 | 0.0546 | No | ||
| 107 | LDHB | 1416183_a_at 1434499_a_at 1442618_at 1448237_x_at 1455235_x_at | 16211 | -0.142 | 0.0239 | No | ||
| 108 | EMD | 1417357_at | 17120 | -0.219 | -0.0158 | No | ||
| 109 | PSME4 | 1426823_s_at 1426824_at 1440511_at 1445147_at 1446464_at 1452211_at | 17227 | -0.230 | -0.0188 | No | ||
| 110 | MMP15 | 1422597_at 1437462_x_at 1439734_at | 17266 | -0.234 | -0.0185 | No | ||
| 111 | PDCD4 | 1444171_at | 17719 | -0.287 | -0.0368 | No | ||
| 112 | POLRMT | 1437377_a_at 1452835_a_at | 18031 | -0.327 | -0.0483 | No | ||
| 113 | CCT4 | 1415867_at 1415868_at 1430034_at 1433447_x_at 1438560_x_at 1442569_at 1456082_x_at 1456572_x_at | 18523 | -0.400 | -0.0675 | No | ||
| 114 | CDC20 | 1416664_at 1439377_x_at 1439394_x_at | 18719 | -0.438 | -0.0727 | No | ||
| 115 | GGA2 | 1428141_at 1457955_at | 19059 | -0.513 | -0.0839 | No | ||
| 116 | TOP1 | 1423474_at 1438495_at 1457573_at 1459672_at | 19089 | -0.518 | -0.0809 | No | ||
| 117 | DYRK1A | 1426226_at 1428299_at 1451977_at | 19263 | -0.563 | -0.0840 | No | ||
| 118 | PTBP1 | 1424874_a_at 1450443_at 1458284_at | 19598 | -0.644 | -0.0939 | No | ||
| 119 | PTPN6 | 1456694_x_at 1460188_at | 19706 | -0.676 | -0.0931 | No | ||
| 120 | SPINT2 | 1438968_x_at 1451935_a_at | 19742 | -0.691 | -0.0889 | No | ||
| 121 | BRD2 | 1423502_at 1437210_a_at | 20248 | -0.866 | -0.1047 | No | ||
| 122 | LIMK2 | 1418581_a_at 1439896_at 1452060_a_at 1458628_at | 20566 | -1.026 | -0.1106 | No | ||
| 123 | WDR1 | 1423054_at 1437591_a_at 1450851_at | 20635 | -1.070 | -0.1046 | No | ||
| 124 | IDI1 | 1423804_a_at 1451122_at | 20848 | -1.216 | -0.1041 | No | ||
| 125 | MTHFD1 | 1415916_a_at 1415917_at 1436704_x_at | 20905 | -1.269 | -0.0959 | No | ||
| 126 | UMPS | 1431652_at 1434859_at 1451445_at 1455832_a_at | 20911 | -1.275 | -0.0854 | No | ||
| 127 | TUBA1 | 1418884_x_at | 21170 | -1.553 | -0.0841 | No | ||
| 128 | TCEB1 | 1427914_a_at 1427915_s_at 1436226_at 1452593_a_at | 21332 | -1.774 | -0.0764 | No | ||
| 129 | TNFRSF1A | 1417291_at | 21427 | -1.934 | -0.0644 | No | ||
| 130 | RASSF2 | 1428392_at 1444889_at | 21668 | -2.567 | -0.0537 | No | ||
| 131 | PSMD14 | 1421751_a_at 1446521_at 1457765_at 1459734_at | 21855 | -3.630 | -0.0315 | No | ||
| 132 | PSMF1 | 1424369_at 1424370_s_at 1424371_at 1451337_at | 21890 | -4.152 | 0.0020 | No |

