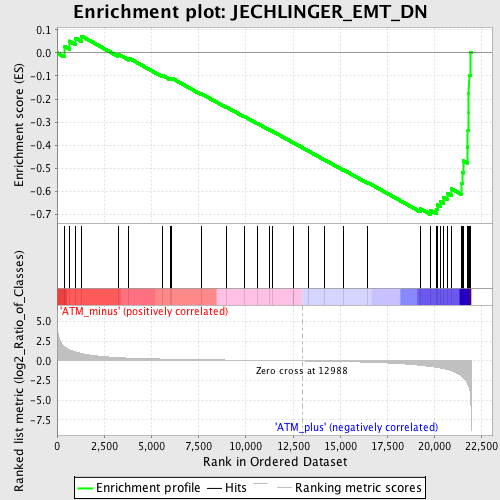
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | JECHLINGER_EMT_DN |
| Enrichment Score (ES) | -0.69979084 |
| Normalized Enrichment Score (NES) | -1.8395737 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.06140921 |
| FWER p-Value | 0.228 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | AMD1 | 1448484_at | 368 | 1.845 | 0.0288 | No | ||
| 2 | MYH9 | 1417472_at 1420170_at 1420171_s_at 1420172_at 1440708_at | 640 | 1.418 | 0.0514 | No | ||
| 3 | ITGB5 | 1417533_a_at 1417534_at 1456133_x_at 1456195_x_at | 984 | 1.131 | 0.0637 | No | ||
| 4 | SERPINB5 | 1421752_a_at 1424623_at 1438856_x_at 1441941_x_at | 1301 | 0.943 | 0.0725 | No | ||
| 5 | ATF3 | 1449363_at | 3225 | 0.420 | -0.0049 | No | ||
| 6 | FOSB | 1422134_at | 3800 | 0.354 | -0.0224 | No | ||
| 7 | ITPR1 | 1417279_at 1443492_at 1457189_at 1460203_at | 5578 | 0.239 | -0.0976 | No | ||
| 8 | CTGF | 1416953_at | 5978 | 0.221 | -0.1104 | No | ||
| 9 | TACSTD1 | 1416579_a_at 1447899_x_at | 6081 | 0.218 | -0.1097 | No | ||
| 10 | CTSH | 1418365_at 1443814_x_at | 7633 | 0.161 | -0.1765 | No | ||
| 11 | TGFB3 | 1417455_at | 8977 | 0.118 | -0.2350 | No | ||
| 12 | TIMP3 | 1419088_at 1419089_at 1449334_at 1449335_at | 9910 | 0.091 | -0.2753 | No | ||
| 13 | VAMP8 | 1420624_a_at | 10599 | 0.071 | -0.3050 | No | ||
| 14 | KITLG | 1415854_at 1415855_at 1426152_a_at 1440621_at 1448117_at | 11223 | 0.054 | -0.3321 | No | ||
| 15 | CDH1 | 1443509_at 1448261_at | 11382 | 0.049 | -0.3381 | No | ||
| 16 | GRB7 | 1448227_at | 12515 | 0.015 | -0.3894 | No | ||
| 17 | FBP2 | 1449088_at | 12537 | 0.014 | -0.3900 | No | ||
| 18 | NUMB | 1416891_at 1425368_a_at 1444337_at 1457175_at | 13287 | -0.010 | -0.4240 | No | ||
| 19 | F3 | 1417408_at | 14164 | -0.042 | -0.4630 | No | ||
| 20 | HMMR | 1425815_a_at 1427541_x_at 1429871_at 1450156_a_at 1450157_a_at | 15168 | -0.085 | -0.5067 | No | ||
| 21 | PKP1 | 1449586_at | 16433 | -0.158 | -0.5605 | No | ||
| 22 | TGM2 | 1417500_a_at 1426004_a_at 1433428_x_at 1437277_x_at 1438942_x_at 1455900_x_at | 19234 | -0.554 | -0.6747 | No | ||
| 23 | STAT5A | 1421469_a_at 1450259_a_at | 19785 | -0.702 | -0.6825 | Yes | ||
| 24 | SGK | 1416041_at | 20087 | -0.813 | -0.6761 | Yes | ||
| 25 | TIAM1 | 1418057_at 1444373_at 1453887_a_at | 20128 | -0.828 | -0.6575 | Yes | ||
| 26 | IRF6 | 1418301_at | 20304 | -0.891 | -0.6435 | Yes | ||
| 27 | JUP | 1426873_s_at | 20476 | -0.969 | -0.6274 | Yes | ||
| 28 | FLNA | 1426677_at | 20659 | -1.088 | -0.6088 | Yes | ||
| 29 | RARA | 1450180_a_at | 20869 | -1.233 | -0.5879 | Yes | ||
| 30 | ACTN4 | 1423449_a_at 1444258_at | 21433 | -1.946 | -0.5656 | Yes | ||
| 31 | ARHGEF1 | 1421164_a_at 1440403_at | 21451 | -1.983 | -0.5174 | Yes | ||
| 32 | DUSP1 | 1448830_at | 21513 | -2.123 | -0.4678 | Yes | ||
| 33 | KLF2 | 1448890_at | 21716 | -2.815 | -0.4075 | Yes | ||
| 34 | ATP1A1 | 1423653_at 1435919_at 1435920_x_at 1451071_a_at 1458868_at | 21758 | -2.992 | -0.3355 | Yes | ||
| 35 | BCL6 | 1421818_at 1450381_a_at | 21807 | -3.243 | -0.2576 | Yes | ||
| 36 | EGR1 | 1417065_at | 21809 | -3.263 | -0.1771 | Yes | ||
| 37 | NNT | 1416105_at 1432608_at 1456573_x_at | 21813 | -3.281 | -0.0962 | Yes | ||
| 38 | EGR2 | 1427682_a_at 1427683_at | 21887 | -4.118 | 0.0021 | Yes |

