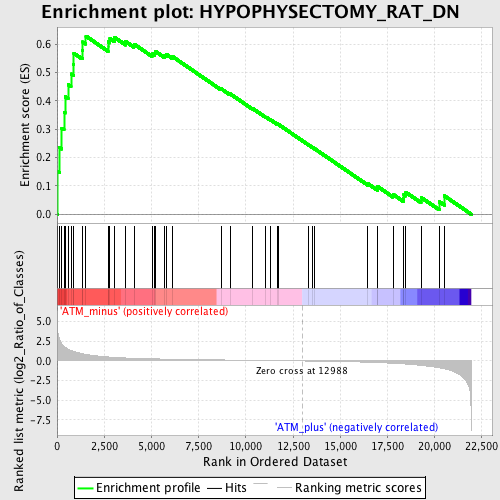
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_minus |
| GeneSet | HYPOPHYSECTOMY_RAT_DN |
| Enrichment Score (ES) | 0.6279262 |
| Normalized Enrichment Score (NES) | 1.7964798 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.050705362 |
| FWER p-Value | 0.509 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | FBP1 | 1448470_at | 15 | 4.587 | 0.1523 | Yes | ||
| 2 | SPARC | 1416589_at 1448392_at 1458204_at | 139 | 2.643 | 0.2349 | Yes | ||
| 3 | HADH | 1436756_x_at 1455972_x_at 1460184_at | 249 | 2.195 | 0.3032 | Yes | ||
| 4 | COL3A1 | 1427883_a_at 1427884_at 1442977_at | 367 | 1.847 | 0.3594 | Yes | ||
| 5 | DECR1 | 1419367_at 1449443_at | 422 | 1.731 | 0.4147 | Yes | ||
| 6 | PIM3 | 1437100_x_at 1438956_x_at 1451069_at | 582 | 1.501 | 0.4575 | Yes | ||
| 7 | TKT | 1436605_at 1439443_x_at 1451015_at | 737 | 1.321 | 0.4945 | Yes | ||
| 8 | ACAA2 | 1428145_at 1428146_s_at 1455061_a_at | 877 | 1.201 | 0.5283 | Yes | ||
| 9 | PDIA3 | 1423423_at | 888 | 1.193 | 0.5676 | Yes | ||
| 10 | FN1 | 1426642_at 1437218_at | 1332 | 0.928 | 0.5783 | Yes | ||
| 11 | MGP | 1448416_at | 1364 | 0.913 | 0.6074 | Yes | ||
| 12 | LDHA | 1419737_a_at | 1527 | 0.839 | 0.6279 | Yes | ||
| 13 | SCD | 1415964_at 1415965_at | 2700 | 0.503 | 0.5912 | No | ||
| 14 | AHSG | 1455093_a_at | 2702 | 0.503 | 0.6079 | No | ||
| 15 | ARF4 | 1423052_at 1423053_at 1434074_x_at 1439367_x_at 1442905_at 1457348_at | 2796 | 0.482 | 0.6197 | No | ||
| 16 | OAT | 1416452_at | 3036 | 0.444 | 0.6236 | No | ||
| 17 | GLUD1 | 1416209_at 1448253_at | 3631 | 0.373 | 0.6089 | No | ||
| 18 | LIMS2 | 1424408_at | 4071 | 0.330 | 0.5999 | No | ||
| 19 | ACSL1 | 1422526_at 1423883_at 1447355_at 1450643_s_at 1460316_at | 5028 | 0.268 | 0.5651 | No | ||
| 20 | ENO1 | 1419023_x_at | 5178 | 0.260 | 0.5670 | No | ||
| 21 | RAB3B | 1422583_at | 5213 | 0.258 | 0.5741 | No | ||
| 22 | B2M | 1427511_at 1449289_a_at 1452428_a_at | 5693 | 0.234 | 0.5600 | No | ||
| 23 | TFF3 | 1417370_at | 5789 | 0.230 | 0.5633 | No | ||
| 24 | FDPS | 1423418_at | 6092 | 0.217 | 0.5568 | No | ||
| 25 | FTH1 | 1448771_a_at | 8680 | 0.127 | 0.4428 | No | ||
| 26 | FBLN5 | 1416164_at 1446377_at | 9158 | 0.113 | 0.4248 | No | ||
| 27 | PSMB8 | 1422962_a_at 1444619_x_at | 10343 | 0.078 | 0.3733 | No | ||
| 28 | CALR | 1417606_a_at 1433806_x_at 1456170_x_at | 11055 | 0.058 | 0.3428 | No | ||
| 29 | ALDOB | 1451194_at | 11301 | 0.051 | 0.3333 | No | ||
| 30 | NPPA | 1456062_at | 11646 | 0.041 | 0.3189 | No | ||
| 31 | DAD1 | 1418528_a_at 1454860_x_at | 11732 | 0.038 | 0.3163 | No | ||
| 32 | PDK4 | 1417273_at | 13298 | -0.011 | 0.2452 | No | ||
| 33 | DDC | 1426215_at 1430591_at 1445524_at | 13525 | -0.018 | 0.2355 | No | ||
| 34 | MEP1B | 1418215_at | 13649 | -0.023 | 0.2306 | No | ||
| 35 | PPGB | 1445659_at 1448128_at 1458466_at | 16460 | -0.160 | 0.1076 | No | ||
| 36 | DCN | 1441506_at 1449368_at | 16968 | -0.203 | 0.0912 | No | ||
| 37 | CYB5B | 1417766_at 1417767_at 1443202_at 1448844_at | 16980 | -0.205 | 0.0975 | No | ||
| 38 | PEX19 | 1416425_at 1442815_at 1448332_at 1455208_at | 17788 | -0.295 | 0.0705 | No | ||
| 39 | AQP8 | 1417828_at | 18350 | -0.372 | 0.0573 | No | ||
| 40 | SLC9A1 | 1417397_at | 18363 | -0.374 | 0.0692 | No | ||
| 41 | ALAS1 | 1424126_at 1442331_at 1455282_x_at | 18470 | -0.390 | 0.0774 | No | ||
| 42 | ECH1 | 1448491_at | 19274 | -0.565 | 0.0595 | No | ||
| 43 | IDH1 | 1419821_s_at 1422433_s_at | 20247 | -0.865 | 0.0440 | No | ||
| 44 | ATP2A2 | 1416551_at 1427250_at 1427251_at 1437797_at 1443551_at 1452363_a_at 1458927_at | 20510 | -0.989 | 0.0650 | No |

