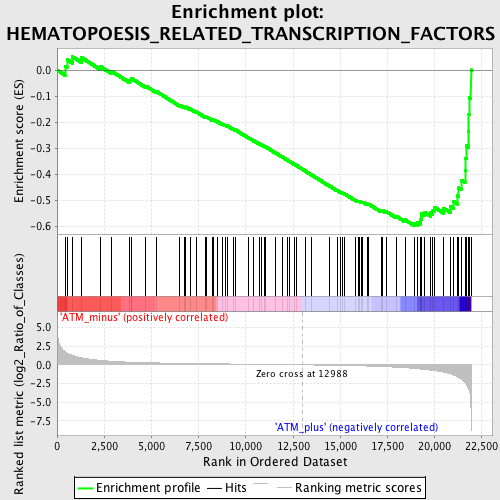
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_02_ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus.phenotype_ATM_minus_versus_ATM_plus.cls #ATM_minus_versus_ATM_plus_repos |
| Phenotype | phenotype_ATM_minus_versus_ATM_plus.cls#ATM_minus_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | HEMATOPOESIS_RELATED_TRANSCRIPTION_FACTORS |
| Enrichment Score (ES) | -0.5976767 |
| Normalized Enrichment Score (NES) | -1.7625628 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.107180536 |
| FWER p-Value | 0.66 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | CEBPB | 1418901_at 1427844_a_at | 421 | 1.731 | 0.0145 | No | ||
| 2 | MYC | 1424942_a_at | 529 | 1.566 | 0.0401 | No | ||
| 3 | NOTCH1 | 1418633_at 1418634_at | 823 | 1.247 | 0.0510 | No | ||
| 4 | HMGB3 | 1416155_at | 1304 | 0.940 | 0.0474 | No | ||
| 5 | PRRX1 | 1425526_a_at 1425527_at 1425528_at 1432129_a_at 1439774_at 1458004_at | 2285 | 0.593 | 0.0141 | No | ||
| 6 | LHX2 | 1418317_at | 2879 | 0.468 | -0.0039 | No | ||
| 7 | HOXD3 | 1421537_at 1437664_at | 3818 | 0.352 | -0.0400 | No | ||
| 8 | TCF3 | 1436207_at 1450117_at | 3912 | 0.345 | -0.0375 | No | ||
| 9 | CEBPA | 1418982_at | 3951 | 0.341 | -0.0326 | No | ||
| 10 | HOXA5 | 1443803_x_at 1448926_at | 4702 | 0.287 | -0.0613 | No | ||
| 11 | NF1 | 1438067_at 1441523_at 1443097_at 1452525_a_at | 5271 | 0.256 | -0.0823 | No | ||
| 12 | HOXC6 | 1427361_at 1427362_x_at 1427454_at | 6502 | 0.200 | -0.1347 | No | ||
| 13 | FOXD3 | 1422210_at | 6732 | 0.192 | -0.1415 | No | ||
| 14 | WT1 | 1425995_s_at 1443221_at | 6807 | 0.189 | -0.1412 | No | ||
| 15 | MEIS1 | 1440431_at 1443260_at 1445773_at 1450992_a_at 1459423_at 1459512_at | 7051 | 0.181 | -0.1488 | No | ||
| 16 | HOXB3 | 1427605_at 1456229_at | 7368 | 0.169 | -0.1599 | No | ||
| 17 | HOXD13 | 1422239_at 1440626_at | 7865 | 0.153 | -0.1796 | No | ||
| 18 | POU2F1 | 1419716_a_at 1427695_a_at 1427835_at | 7929 | 0.151 | -0.1796 | No | ||
| 19 | SPI1 | 1418747_at | 8242 | 0.141 | -0.1911 | No | ||
| 20 | NFIB | 1416293_at 1427680_a_at 1434101_at 1434102_at 1438072_at 1438244_at 1442214_at 1444800_at 1448288_at 1454834_at | 8297 | 0.140 | -0.1908 | No | ||
| 21 | PAX6 | 1419271_at 1425960_s_at 1437816_at 1452526_a_at 1456342_at | 8486 | 0.134 | -0.1968 | No | ||
| 22 | HOXB6 | 1451660_a_at | 8772 | 0.124 | -0.2074 | No | ||
| 23 | KLF1 | 1418600_at | 8892 | 0.121 | -0.2105 | No | ||
| 24 | SRF | 1418255_s_at 1418256_at | 9000 | 0.117 | -0.2131 | No | ||
| 25 | CREBBP | 1434633_at 1435224_at 1436983_at 1444856_at 1457641_at 1459804_at | 9321 | 0.108 | -0.2257 | No | ||
| 26 | HOXA7 | 1449499_at | 9428 | 0.105 | -0.2285 | No | ||
| 27 | HIF1A | 1427418_a_at 1431981_at 1448183_a_at 1457231_at | 10124 | 0.084 | -0.2587 | No | ||
| 28 | HHEX | 1423319_at | 10424 | 0.076 | -0.2709 | No | ||
| 29 | GATA1 | 1449232_at | 10714 | 0.067 | -0.2828 | No | ||
| 30 | NFIX | 1423493_a_at 1436363_a_at 1436364_x_at 1451443_at 1459909_at | 10848 | 0.063 | -0.2876 | No | ||
| 31 | ETV6 | 1423401_at 1434880_at 1444598_at 1457944_at | 10973 | 0.060 | -0.2921 | No | ||
| 32 | GATA2 | 1428816_a_at 1450333_a_at | 11018 | 0.059 | -0.2930 | No | ||
| 33 | USF2 | 1418241_at 1460228_at | 11539 | 0.044 | -0.3159 | No | ||
| 34 | NFKB1 | 1427705_a_at 1442949_at | 11951 | 0.032 | -0.3341 | No | ||
| 35 | BMP4 | 1422912_at | 12215 | 0.024 | -0.3457 | No | ||
| 36 | HLX1 | 1418939_at | 12295 | 0.021 | -0.3489 | No | ||
| 37 | HOXA10 | 1431475_a_at 1446408_at | 12570 | 0.013 | -0.3612 | No | ||
| 38 | HOXB8 | 1452493_s_at | 12677 | 0.010 | -0.3658 | No | ||
| 39 | POU1F1 | 1422220_at | 13142 | -0.005 | -0.3869 | No | ||
| 40 | PAX8 | 1418208_at 1446561_at | 13451 | -0.016 | -0.4007 | No | ||
| 41 | PBX1 | 1430183_at 1440037_at 1440954_at 1442846_at 1449542_at 1458853_at | 14435 | -0.052 | -0.4447 | No | ||
| 42 | LMO2 | 1454086_a_at | 14833 | -0.068 | -0.4615 | No | ||
| 43 | EPAS1 | 1435436_at 1449888_at | 15005 | -0.076 | -0.4679 | No | ||
| 44 | EED | 1448653_at | 15112 | -0.082 | -0.4711 | No | ||
| 45 | NFIC | 1422565_s_at 1427665_a_at 1429148_at 1435527_at 1440140_at 1450661_x_at | 15227 | -0.087 | -0.4746 | No | ||
| 46 | ETV4 | 1423232_at 1443381_at | 15791 | -0.115 | -0.4982 | No | ||
| 47 | GBX2 | 1420337_at | 15952 | -0.125 | -0.5030 | No | ||
| 48 | TAL1 | 1449389_at | 15993 | -0.127 | -0.5024 | No | ||
| 49 | GATA3 | 1448886_at | 16129 | -0.136 | -0.5059 | No | ||
| 50 | DLX4 | 1420595_at 1427641_at 1443591_at | 16192 | -0.141 | -0.5060 | No | ||
| 51 | JUN | 1417409_at 1448694_at | 16418 | -0.157 | -0.5132 | No | ||
| 52 | MEFV | 1460283_at | 16506 | -0.163 | -0.5140 | No | ||
| 53 | NFYA | 1422082_a_at 1427808_at 1452560_a_at | 17163 | -0.224 | -0.5397 | No | ||
| 54 | MAFK | 1418616_at | 17207 | -0.228 | -0.5372 | No | ||
| 55 | MAFB | 1451715_at 1451716_at | 17440 | -0.254 | -0.5429 | No | ||
| 56 | NFE2 | 1452001_at | 17962 | -0.320 | -0.5605 | No | ||
| 57 | SP3 | 1431804_a_at 1459360_at | 18430 | -0.383 | -0.5744 | No | ||
| 58 | E2F1 | 1417878_at 1431875_a_at 1447840_x_at | 18940 | -0.485 | -0.5882 | Yes | ||
| 59 | NFKB2 | 1425902_a_at 1429128_x_at | 19072 | -0.515 | -0.5842 | Yes | ||
| 60 | IRF1 | 1448436_a_at | 19248 | -0.560 | -0.5813 | Yes | ||
| 61 | PHC1 | 1423212_at | 19267 | -0.564 | -0.5711 | Yes | ||
| 62 | PAX5 | 1421765_at 1459916_at | 19281 | -0.568 | -0.5606 | Yes | ||
| 63 | EP300 | 1434765_at | 19298 | -0.572 | -0.5502 | Yes | ||
| 64 | BMI1 | 1417493_at 1448733_at | 19470 | -0.611 | -0.5461 | Yes | ||
| 65 | STAT5A | 1421469_a_at 1450259_a_at | 19785 | -0.702 | -0.5468 | Yes | ||
| 66 | KLF3 | 1421604_a_at 1429360_at 1431630_a_at 1441200_at 1442920_at 1454666_at | 19902 | -0.745 | -0.5376 | Yes | ||
| 67 | MYB | 1421317_x_at 1422734_a_at 1450194_a_at | 19972 | -0.769 | -0.5257 | Yes | ||
| 68 | ELF1 | 1417540_at 1439319_at | 20470 | -0.965 | -0.5297 | Yes | ||
| 69 | GFI1 | 1417679_at | 20817 | -1.196 | -0.5222 | Yes | ||
| 70 | FLI1 | 1422024_at 1433512_at 1441584_at | 21006 | -1.360 | -0.5043 | Yes | ||
| 71 | E2F2 | 1455790_at 1458574_at | 21210 | -1.602 | -0.4823 | Yes | ||
| 72 | HOXA9 | 1421579_at 1455626_at | 21257 | -1.668 | -0.4519 | Yes | ||
| 73 | KLF13 | 1432543_a_at 1456030_at | 21412 | -1.910 | -0.4217 | Yes | ||
| 74 | ETS1 | 1422027_a_at 1422028_a_at 1426725_s_at 1452163_at | 21604 | -2.342 | -0.3848 | Yes | ||
| 75 | XBP1 | 1420011_s_at 1420012_at 1420886_a_at 1437223_s_at | 21644 | -2.460 | -0.3386 | Yes | ||
| 76 | RUNX1 | 1422864_at 1422865_at 1427650_a_at 1427847_at 1440878_at 1444527_at 1446930_at 1452530_a_at 1452531_at 1452578_at 1459470_at | 21665 | -2.559 | -0.2897 | Yes | ||
| 77 | STAT1 | 1420915_at 1440481_at 1450033_a_at 1450034_at | 21799 | -3.193 | -0.2335 | Yes | ||
| 78 | EGR1 | 1417065_at | 21809 | -3.263 | -0.1703 | Yes | ||
| 79 | STAT3 | 1424272_at 1426587_a_at 1459961_a_at 1460700_at | 21823 | -3.342 | -0.1058 | Yes | ||
| 80 | ETS2 | 1416268_at 1437809_x_at 1447685_x_at | 21919 | -5.684 | 0.0007 | Yes |

