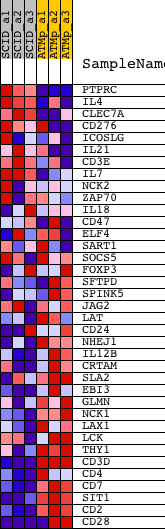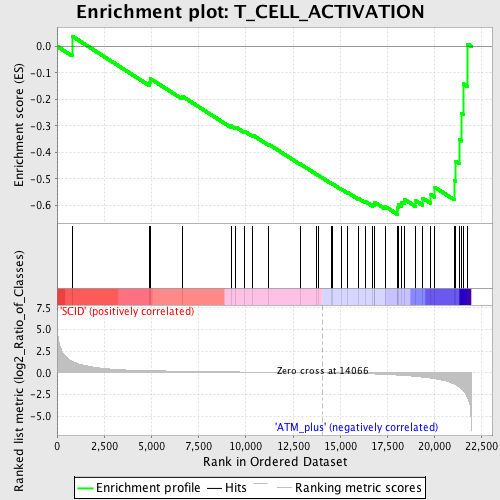
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus_repos |
| Phenotype | phenotype_SCID_versus_ATM_plus.cls#SCID_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | T_CELL_ACTIVATION |
| Enrichment Score (ES) | -0.63423705 |
| Normalized Enrichment Score (NES) | -1.599007 |
| Nominal p-value | 0.013972056 |
| FDR q-value | 0.39034078 |
| FWER p-Value | 1.0 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | PTPRC | 1422124_a_at 1440165_at | 795 | 1.337 | 0.0386 | No | ||
| 2 | IL4 | 1449864_at | 4912 | 0.257 | -0.1350 | No | ||
| 3 | CLEC7A | 1420699_at | 4946 | 0.255 | -0.1222 | No | ||
| 4 | CD276 | 1417599_at | 6617 | 0.190 | -0.1878 | No | ||
| 5 | ICOSLG | 1419212_at 1426087_at | 9209 | 0.119 | -0.2995 | No | ||
| 6 | IL21 | 1450334_at | 9461 | 0.113 | -0.3046 | No | ||
| 7 | CD3E | 1422105_at 1445748_at | 9934 | 0.101 | -0.3205 | No | ||
| 8 | IL7 | 1422080_at 1436861_at | 10371 | 0.091 | -0.3353 | No | ||
| 9 | NCK2 | 1416796_at 1416797_at 1458432_at | 11200 | 0.072 | -0.3691 | No | ||
| 10 | ZAP70 | 1422701_at 1439749_at 1440178_x_at | 12867 | 0.033 | -0.4433 | No | ||
| 11 | IL18 | 1417932_at | 13729 | 0.009 | -0.4821 | No | ||
| 12 | CD47 | 1419554_at 1428187_at 1430997_at 1449507_a_at 1458634_at 1459908_at | 13862 | 0.005 | -0.4878 | No | ||
| 13 | ELF4 | 1421337_at 1445778_at | 14548 | -0.015 | -0.5183 | No | ||
| 14 | SART1 | 1448658_at | 14591 | -0.016 | -0.5192 | No | ||
| 15 | SOCS5 | 1423349_at 1423350_at 1441640_at 1442890_at | 15045 | -0.031 | -0.5382 | No | ||
| 16 | FOXP3 | 1420765_a_at 1457082_at | 15403 | -0.045 | -0.5520 | No | ||
| 17 | SFTPD | 1420378_at | 15938 | -0.067 | -0.5726 | No | ||
| 18 | SPINK5 | 1430567_at | 16315 | -0.086 | -0.5849 | No | ||
| 19 | JAG2 | 1426430_at 1426431_at | 16725 | -0.114 | -0.5973 | No | ||
| 20 | LAT | 1460651_at | 16809 | -0.119 | -0.5944 | No | ||
| 21 | CD24 | 1416034_at 1437502_x_at 1448182_a_at | 16822 | -0.120 | -0.5882 | No | ||
| 22 | NHEJ1 | 1425651_at 1429098_s_at | 17369 | -0.167 | -0.6038 | No | ||
| 23 | IL12B | 1419530_at 1449497_at | 18037 | -0.244 | -0.6206 | Yes | ||
| 24 | CRTAM | 1449903_at | 18041 | -0.245 | -0.6070 | Yes | ||
| 25 | SLA2 | 1437504_at 1451492_at | 18091 | -0.251 | -0.5952 | Yes | ||
| 26 | EBI3 | 1449222_at | 18256 | -0.275 | -0.5872 | Yes | ||
| 27 | GLMN | 1418839_at | 18398 | -0.294 | -0.5772 | Yes | ||
| 28 | NCK1 | 1421487_a_at 1424543_at 1447271_at | 18972 | -0.389 | -0.5816 | Yes | ||
| 29 | LAX1 | 1438687_at | 19357 | -0.475 | -0.5725 | Yes | ||
| 30 | LCK | 1425396_a_at 1439145_at 1439146_s_at 1457917_at | 19798 | -0.593 | -0.5594 | Yes | ||
| 31 | THY1 | 1423135_at | 20009 | -0.662 | -0.5319 | Yes | ||
| 32 | CD3D | 1422828_at | 21050 | -1.299 | -0.5067 | Yes | ||
| 33 | CD4 | 1419696_at 1427779_a_at | 21103 | -1.360 | -0.4328 | Yes | ||
| 34 | CD7 | 1419711_at | 21291 | -1.612 | -0.3511 | Yes | ||
| 35 | SIT1 | 1418751_at | 21424 | -1.851 | -0.2534 | Yes | ||
| 36 | CD2 | 1418770_at | 21523 | -2.075 | -0.1417 | Yes | ||
| 37 | CD28 | 1417597_at 1437025_at 1443703_at | 21746 | -2.866 | 0.0087 | Yes |