Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus_repos |
| Phenotype | phenotype_SCID_versus_ATM_plus.cls#SCID_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
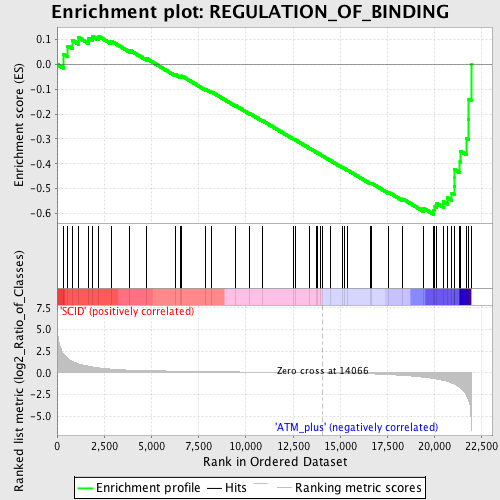
| GeneSet | REGULATION_OF_BINDING |
| Enrichment Score (ES) | -0.60369384 |
| Normalized Enrichment Score (NES) | -1.6454717 |
| Nominal p-value | 0.003937008 |
| FDR q-value | 0.2935851 |
| FWER p-Value | 0.983 |

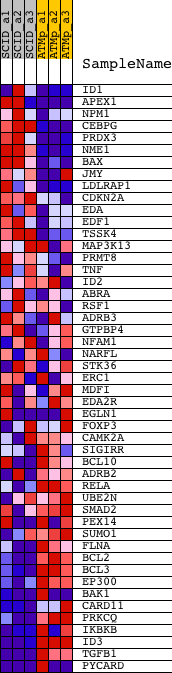
| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ID1 | 1425895_a_at | 358 | 2.138 | 0.0392 | No | ||
| 2 | APEX1 | 1416135_at 1437715_x_at 1456079_x_at | 567 | 1.683 | 0.0734 | No | ||
| 3 | NPM1 | 1415839_a_at 1420267_at 1420268_x_at 1420269_at 1432416_a_at | 828 | 1.306 | 0.0955 | No | ||
| 4 | CEBPG | 1425261_at 1425262_at 1451639_at | 1142 | 1.027 | 0.1079 | No | ||
| 5 | PRDX3 | 1416292_at | 1646 | 0.768 | 0.1048 | No | ||
| 6 | NME1 | 1424110_a_at 1435277_x_at | 1875 | 0.678 | 0.1120 | No | ||
| 7 | BAX | 1416837_at | 2180 | 0.583 | 0.1133 | No | ||
| 8 | JMY | 1420639_at 1420640_at 1451878_a_at | 2862 | 0.441 | 0.0936 | No | ||
| 9 | LDLRAP1 | 1424378_at 1457162_at | 3846 | 0.321 | 0.0571 | No | ||
| 10 | CDKN2A | 1450140_a_at | 4728 | 0.265 | 0.0237 | No | ||
| 11 | EDA | 1419597_at 1449524_at 1451877_at 1451925_at | 6256 | 0.203 | -0.0408 | No | ||
| 12 | EDF1 | 1440151_s_at 1448266_at | 6521 | 0.194 | -0.0478 | No | ||
| 13 | TSSK4 | 1432045_at 1432101_a_at 1438486_at 1443810_at | 6612 | 0.191 | -0.0470 | No | ||
| 14 | MAP3K13 | 1430137_at 1440749_at 1447699_at | 7862 | 0.153 | -0.1001 | No | ||
| 15 | PRMT8 | 1435204_at | 8189 | 0.144 | -0.1112 | No | ||
| 16 | TNF | 1419607_at | 9455 | 0.113 | -0.1661 | No | ||
| 17 | ID2 | 1422537_a_at 1435176_a_at 1453596_at | 10203 | 0.095 | -0.1977 | No | ||
| 18 | ABRA | 1458455_at | 10895 | 0.079 | -0.2273 | No | ||
| 19 | RSF1 | 1438735_at | 12537 | 0.040 | -0.3012 | No | ||
| 20 | ADRB3 | 1421555_at 1455918_at | 12618 | 0.038 | -0.3038 | No | ||
| 21 | GTPBP4 | 1423142_a_at 1423143_at 1450873_at | 13357 | 0.019 | -0.3371 | No | ||
| 22 | NFAM1 | 1425714_a_at 1428790_at | 13722 | 0.009 | -0.3535 | No | ||
| 23 | NARFL | 1450452_a_at | 13809 | 0.007 | -0.3572 | No | ||
| 24 | STK36 | 1434733_at 1453584_at | 13949 | 0.003 | -0.3635 | No | ||
| 25 | ERC1 | 1421501_a_at 1424247_at 1441493_at 1444356_at 1451279_at | 14036 | 0.001 | -0.3674 | No | ||
| 26 | MDFI | 1420713_a_at 1425924_at | 14488 | -0.013 | -0.3876 | No | ||
| 27 | EDA2R | 1440085_at | 15099 | -0.033 | -0.4147 | No | ||
| 28 | EGLN1 | 1423785_at 1443389_at 1451110_at | 15218 | -0.038 | -0.4191 | No | ||
| 29 | FOXP3 | 1420765_a_at 1457082_at | 15403 | -0.045 | -0.4263 | No | ||
| 30 | CAMK2A | 1437125_at 1441734_at 1442707_at 1443876_at 1452453_a_at 1457311_at | 16595 | -0.105 | -0.4780 | No | ||
| 31 | SIGIRR | 1449163_at | 16659 | -0.109 | -0.4780 | No | ||
| 32 | BCL10 | 1418970_a_at 1418971_x_at 1418972_at 1443524_x_at | 17549 | -0.185 | -0.5139 | No | ||
| 33 | ADRB2 | 1437302_at | 18314 | -0.283 | -0.5414 | No | ||
| 34 | RELA | 1419536_a_at | 19403 | -0.485 | -0.5785 | No | ||
| 35 | UBE2N | 1422559_at 1435384_at | 19955 | -0.642 | -0.5870 | Yes | ||
| 36 | SMAD2 | 1420634_a_at 1458304_at | 20011 | -0.663 | -0.5723 | Yes | ||
| 37 | PEX14 | 1419053_at 1458153_at | 20094 | -0.696 | -0.5580 | Yes | ||
| 38 | SUMO1 | 1438289_a_at 1444557_at 1451005_at 1456349_x_at | 20469 | -0.858 | -0.5528 | Yes | ||
| 39 | FLNA | 1426677_at | 20668 | -0.968 | -0.5366 | Yes | ||
| 40 | BCL2 | 1422938_at 1427818_at 1437122_at 1440770_at 1443837_x_at 1457687_at | 20911 | -1.157 | -0.5176 | Yes | ||
| 41 | BCL3 | 1418133_at | 21029 | -1.282 | -0.4897 | Yes | ||
| 42 | EP300 | 1434765_at | 21052 | -1.301 | -0.4569 | Yes | ||
| 43 | BAK1 | 1418991_at 1425716_s_at | 21055 | -1.304 | -0.4231 | Yes | ||
| 44 | CARD11 | 1430257_at 1435996_at | 21337 | -1.678 | -0.3923 | Yes | ||
| 45 | PRKCQ | 1426044_a_at | 21352 | -1.700 | -0.3488 | Yes | ||
| 46 | IKBKB | 1426207_at 1426333_a_at 1432275_at 1445141_at 1454184_a_at | 21683 | -2.583 | -0.2968 | Yes | ||
| 47 | ID3 | 1416630_at | 21780 | -3.031 | -0.2224 | Yes | ||
| 48 | TGFB1 | 1420653_at 1445360_at | 21809 | -3.227 | -0.1399 | Yes | ||
| 49 | PYCARD | 1417346_at 1417347_at 1446926_at | 21930 | -5.604 | 0.0003 | Yes |