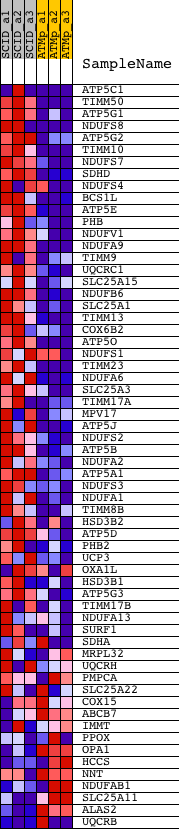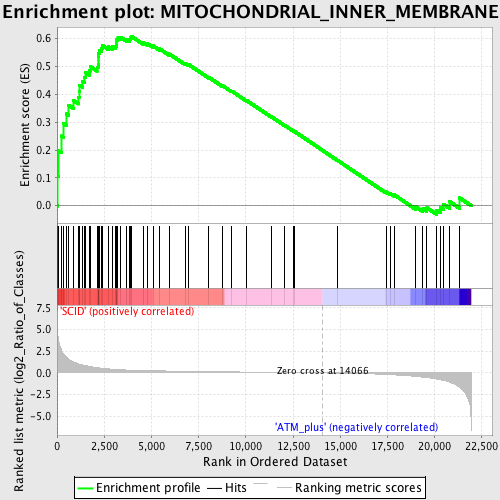
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus_repos |
| Phenotype | phenotype_SCID_versus_ATM_plus.cls#SCID_versus_ATM_plus_repos |
| Upregulated in class | SCID |
| GeneSet | MITOCHONDRIAL_INNER_MEMBRANE |
| Enrichment Score (ES) | 0.6089832 |
| Normalized Enrichment Score (NES) | 1.696071 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.12239248 |
| FWER p-Value | 0.856 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | ATP5C1 | 1416058_s_at 1438809_at | 29 | 4.740 | 0.1067 | Yes | ||
| 2 | TIMM50 | 1441819_x_at 1455456_a_at | 56 | 4.053 | 0.1980 | Yes | ||
| 3 | ATP5G1 | 1416020_a_at 1444874_at | 227 | 2.644 | 0.2505 | Yes | ||
| 4 | NDUFS8 | 1423907_a_at 1423908_at 1434212_at 1434213_x_at 1434579_x_at 1455283_x_at | 337 | 2.196 | 0.2956 | Yes | ||
| 5 | ATP5G2 | 1415980_at 1456128_at | 502 | 1.824 | 0.3297 | Yes | ||
| 6 | TIMM10 | 1417499_at | 620 | 1.571 | 0.3601 | Yes | ||
| 7 | NDUFS7 | 1424313_a_at 1451312_at | 864 | 1.252 | 0.3776 | Yes | ||
| 8 | SDHD | 1428235_at 1437489_x_at | 1135 | 1.031 | 0.3887 | Yes | ||
| 9 | NDUFS4 | 1418117_at 1438166_x_at 1444464_at 1448959_at 1459760_at | 1166 | 1.014 | 0.4105 | Yes | ||
| 10 | BCS1L | 1451541_at | 1183 | 1.005 | 0.4327 | Yes | ||
| 11 | ATP5E | 1416567_s_at | 1345 | 0.915 | 0.4462 | Yes | ||
| 12 | PHB | 1435144_at 1442233_at 1448563_at | 1432 | 0.872 | 0.4621 | Yes | ||
| 13 | NDUFV1 | 1415966_a_at 1415967_at 1456015_x_at | 1496 | 0.842 | 0.4784 | Yes | ||
| 14 | NDUFA9 | 1416663_at | 1706 | 0.737 | 0.4857 | Yes | ||
| 15 | TIMM9 | 1428010_at 1449886_a_at | 1753 | 0.719 | 0.5000 | Yes | ||
| 16 | UQCRC1 | 1428782_a_at | 2115 | 0.601 | 0.4972 | Yes | ||
| 17 | SLC25A15 | 1420966_at 1420967_at | 2173 | 0.585 | 0.5079 | Yes | ||
| 18 | NDUFB6 | 1434056_a_at 1434057_at | 2181 | 0.583 | 0.5209 | Yes | ||
| 19 | SLC25A1 | 1428190_at | 2183 | 0.583 | 0.5341 | Yes | ||
| 20 | TIMM13 | 1455211_a_at | 2188 | 0.582 | 0.5472 | Yes | ||
| 21 | COX6B2 | 1435275_at | 2237 | 0.571 | 0.5581 | Yes | ||
| 22 | ATP5O | 1416278_a_at | 2361 | 0.538 | 0.5647 | Yes | ||
| 23 | NDUFS1 | 1425143_a_at 1453354_at | 2398 | 0.531 | 0.5752 | Yes | ||
| 24 | TIMM23 | 1416485_at | 2711 | 0.466 | 0.5715 | Yes | ||
| 25 | NDUFA6 | 1448427_at | 2922 | 0.431 | 0.5717 | Yes | ||
| 26 | SLC25A3 | 1416300_a_at 1440302_at | 3105 | 0.401 | 0.5726 | Yes | ||
| 27 | TIMM17A | 1426256_at | 3144 | 0.396 | 0.5799 | Yes | ||
| 28 | MPV17 | 1420387_at | 3155 | 0.394 | 0.5884 | Yes | ||
| 29 | ATP5J | 1416143_at | 3166 | 0.393 | 0.5969 | Yes | ||
| 30 | NDUFS2 | 1451096_at | 3223 | 0.385 | 0.6031 | Yes | ||
| 31 | ATP5B | 1416829_at | 3364 | 0.368 | 0.6051 | Yes | ||
| 32 | NDUFA2 | 1417368_s_at | 3685 | 0.334 | 0.5981 | Yes | ||
| 33 | ATP5A1 | 1420037_at 1423111_at 1449710_s_at | 3857 | 0.320 | 0.5976 | Yes | ||
| 34 | NDUFS3 | 1423737_at | 3907 | 0.316 | 0.6025 | Yes | ||
| 35 | NDUFA1 | 1422241_a_at | 3924 | 0.315 | 0.6090 | Yes | ||
| 36 | TIMM8B | 1431665_a_at | 4571 | 0.272 | 0.5857 | No | ||
| 37 | HSD3B2 | 1425127_at | 4763 | 0.264 | 0.5829 | No | ||
| 38 | ATP5D | 1423716_s_at 1456580_s_at | 5080 | 0.249 | 0.5742 | No | ||
| 39 | PHB2 | 1416202_at 1438938_x_at 1455713_x_at 1460035_at | 5434 | 0.234 | 0.5634 | No | ||
| 40 | UCP3 | 1420657_at 1420658_at | 5947 | 0.214 | 0.5448 | No | ||
| 41 | OXA1L | 1423738_at 1451088_a_at 1456845_at | 6784 | 0.185 | 0.5109 | No | ||
| 42 | HSD3B1 | 1448453_at | 6957 | 0.180 | 0.5071 | No | ||
| 43 | ATP5G3 | 1454661_at | 8034 | 0.148 | 0.4613 | No | ||
| 44 | TIMM17B | 1438656_x_at 1460685_at | 8742 | 0.131 | 0.4319 | No | ||
| 45 | NDUFA13 | 1430713_s_at | 9251 | 0.118 | 0.4114 | No | ||
| 46 | SURF1 | 1450561_a_at | 10054 | 0.098 | 0.3770 | No | ||
| 47 | SDHA | 1426688_at 1426689_s_at 1445317_at | 11345 | 0.069 | 0.3196 | No | ||
| 48 | MRPL32 | 1424372_at | 12025 | 0.051 | 0.2897 | No | ||
| 49 | UQCRH | 1452133_at 1453229_s_at | 12494 | 0.041 | 0.2693 | No | ||
| 50 | PMPCA | 1424661_at 1424662_at | 12595 | 0.039 | 0.2656 | No | ||
| 51 | SLC25A22 | 1452653_at | 14844 | -0.024 | 0.1634 | No | ||
| 52 | COX15 | 1426693_x_at 1437982_x_at 1452146_a_at 1460376_a_at | 17422 | -0.173 | 0.0495 | No | ||
| 53 | ABCB7 | 1419931_at 1427490_at 1435006_s_at | 17644 | -0.197 | 0.0439 | No | ||
| 54 | IMMT | 1429533_at 1429534_a_at 1433470_a_at 1437126_at | 17847 | -0.217 | 0.0396 | No | ||
| 55 | PPOX | 1416618_at 1441862_at 1448065_at | 18982 | -0.392 | -0.0033 | No | ||
| 56 | OPA1 | 1418768_at 1434890_at 1449214_a_at | 19377 | -0.480 | -0.0104 | No | ||
| 57 | HCCS | 1420889_at 1420890_at | 19588 | -0.534 | -0.0078 | No | ||
| 58 | NNT | 1416105_at 1432608_at 1456573_x_at | 20107 | -0.700 | -0.0155 | No | ||
| 59 | NDUFAB1 | 1428159_s_at 1428160_at 1435934_at 1447919_x_at 1453565_at 1458678_at | 20293 | -0.778 | -0.0062 | No | ||
| 60 | SLC25A11 | 1426586_at | 20475 | -0.859 | 0.0051 | No | ||
| 61 | ALAS2 | 1451675_a_at | 20784 | -1.052 | 0.0150 | No | ||
| 62 | UQCRB | 1416337_at 1455997_a_at | 21322 | -1.651 | 0.0281 | No |