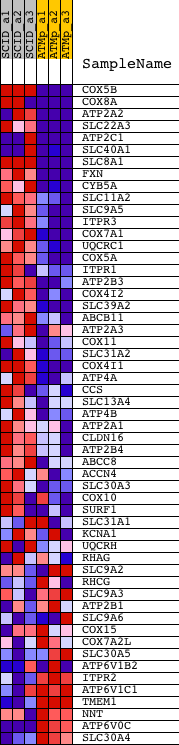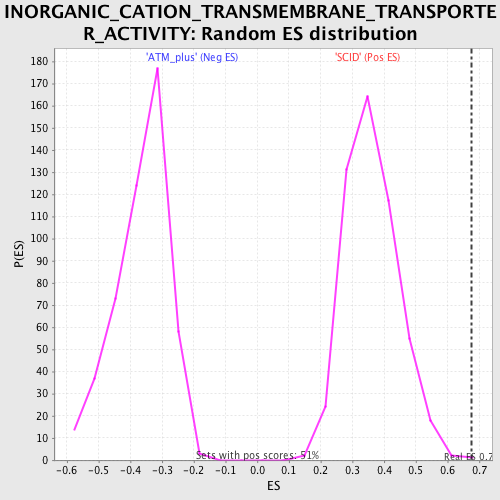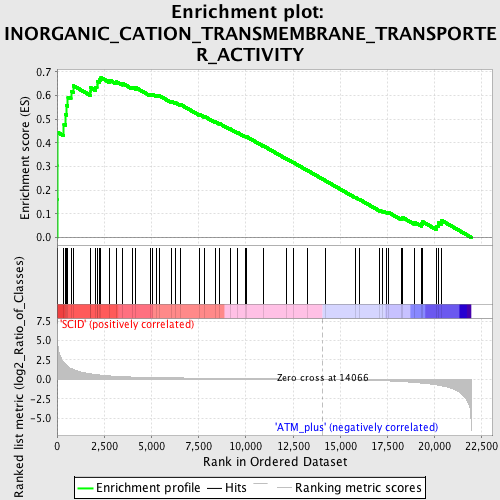
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus_repos |
| Phenotype | phenotype_SCID_versus_ATM_plus.cls#SCID_versus_ATM_plus_repos |
| Upregulated in class | SCID |
| GeneSet | INORGANIC_CATION_TRANSMEMBRANE_TRANSPORTER_ACTIVITY |
| Enrichment Score (ES) | 0.67605245 |
| Normalized Enrichment Score (NES) | 1.8684142 |
| Nominal p-value | 0.0019455253 |
| FDR q-value | 0.040472865 |
| FWER p-Value | 0.072 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | COX5B | 1416902_a_at 1435613_x_at 1436149_at 1448514_at 1454716_x_at 1456588_x_at | 1 | 6.901 | 0.1586 | Yes | ||
| 2 | COX8A | 1416112_at 1436409_at 1448222_x_at | 7 | 6.333 | 0.3040 | Yes | ||
| 3 | ATP2A2 | 1416551_at 1427250_at 1427251_at 1437797_at 1443551_at 1452363_a_at 1458927_at | 9 | 6.169 | 0.4458 | Yes | ||
| 4 | SLC22A3 | 1420444_at | 361 | 2.135 | 0.4789 | Yes | ||
| 5 | ATP2C1 | 1431196_at 1434386_at 1437738_at 1439215_at 1442742_at | 452 | 1.932 | 0.5192 | Yes | ||
| 6 | SLC40A1 | 1417061_at 1447227_at 1448566_at | 510 | 1.804 | 0.5581 | Yes | ||
| 7 | SLC8A1 | 1425817_a_at 1437675_at 1444985_at 1447231_at | 574 | 1.674 | 0.5937 | Yes | ||
| 8 | FXN | 1427282_a_at | 737 | 1.409 | 0.6187 | Yes | ||
| 9 | CYB5A | 1416727_a_at | 870 | 1.245 | 0.6413 | Yes | ||
| 10 | SLC11A2 | 1417584_at 1426441_at 1441709_at 1447333_at 1452078_a_at | 1754 | 0.719 | 0.6175 | Yes | ||
| 11 | SLC9A5 | 1420070_a_at 1455760_at | 1759 | 0.718 | 0.6338 | Yes | ||
| 12 | ITPR3 | 1417297_at | 2034 | 0.625 | 0.6357 | Yes | ||
| 13 | COX7A1 | 1418709_at | 2114 | 0.603 | 0.6459 | Yes | ||
| 14 | UQCRC1 | 1428782_a_at | 2115 | 0.601 | 0.6598 | Yes | ||
| 15 | COX5A | 1415933_a_at 1439267_x_at 1448153_at | 2259 | 0.565 | 0.6662 | Yes | ||
| 16 | ITPR1 | 1417279_at 1443492_at 1457189_at 1460203_at | 2320 | 0.547 | 0.6761 | Yes | ||
| 17 | ATP2B3 | 1442645_at 1445679_at | 2785 | 0.455 | 0.6653 | No | ||
| 18 | COX4I2 | 1421373_at | 3142 | 0.396 | 0.6581 | No | ||
| 19 | SLC39A2 | 1443163_at | 3440 | 0.358 | 0.6528 | No | ||
| 20 | ABCB11 | 1449817_at | 3976 | 0.310 | 0.6355 | No | ||
| 21 | ATP2A3 | 1421129_a_at 1450124_a_at | 4169 | 0.296 | 0.6335 | No | ||
| 22 | COX11 | 1429188_at 1457846_at | 4930 | 0.256 | 0.6047 | No | ||
| 23 | SLC31A2 | 1416654_at 1453721_a_at 1459399_at | 5031 | 0.251 | 0.6059 | No | ||
| 24 | COX4I1 | 1448322_a_at | 5260 | 0.241 | 0.6010 | No | ||
| 25 | ATP4A | 1421286_a_at | 5415 | 0.235 | 0.5994 | No | ||
| 26 | CCS | 1448615_at | 6053 | 0.210 | 0.5751 | No | ||
| 27 | SLC13A4 | 1433734_at | 6278 | 0.202 | 0.5695 | No | ||
| 28 | ATP4B | 1448911_at | 6556 | 0.193 | 0.5613 | No | ||
| 29 | ATP2A1 | 1419312_at | 7545 | 0.163 | 0.5199 | No | ||
| 30 | CLDN16 | 1420434_at | 7831 | 0.153 | 0.5104 | No | ||
| 31 | ATP2B4 | 1445701_at | 8375 | 0.140 | 0.4887 | No | ||
| 32 | ABCC8 | 1455765_a_at 1457066_at | 8577 | 0.135 | 0.4827 | No | ||
| 33 | ACCN4 | 1437091_at | 9156 | 0.120 | 0.4590 | No | ||
| 34 | SLC30A3 | 1460654_at | 9579 | 0.110 | 0.4423 | No | ||
| 35 | COX10 | 1429329_at 1446223_at 1459977_x_at | 9962 | 0.101 | 0.4271 | No | ||
| 36 | SURF1 | 1450561_a_at | 10054 | 0.098 | 0.4252 | No | ||
| 37 | SLC31A1 | 1433750_at 1455285_at | 10920 | 0.078 | 0.3875 | No | ||
| 38 | KCNA1 | 1417416_at 1437230_at 1442413_at 1455785_at | 12135 | 0.049 | 0.3331 | No | ||
| 39 | UQCRH | 1452133_at 1453229_s_at | 12494 | 0.041 | 0.3177 | No | ||
| 40 | RHAG | 1419014_at | 13256 | 0.022 | 0.2834 | No | ||
| 41 | SLC9A2 | 1437259_at | 14213 | -0.004 | 0.2399 | No | ||
| 42 | RHCG | 1417362_at | 15782 | -0.060 | 0.1696 | No | ||
| 43 | SLC9A3 | 1432771_at 1441236_at | 16022 | -0.072 | 0.1603 | No | ||
| 44 | ATP2B1 | 1428936_at 1428937_at 1452969_at 1458346_x_at | 17100 | -0.142 | 0.1144 | No | ||
| 45 | SLC9A6 | 1435008_at 1435009_at | 17252 | -0.156 | 0.1111 | No | ||
| 46 | COX15 | 1426693_x_at 1437982_x_at 1452146_a_at 1460376_a_at | 17422 | -0.173 | 0.1073 | No | ||
| 47 | COX7A2L | 1421772_a_at 1432263_a_at 1432264_x_at | 17553 | -0.185 | 0.1056 | No | ||
| 48 | SLC30A5 | 1422497_at | 18217 | -0.268 | 0.0815 | No | ||
| 49 | ATP6V1B2 | 1415814_at 1419883_s_at 1446465_at 1449649_at | 18299 | -0.281 | 0.0843 | No | ||
| 50 | ITPR2 | 1421678_at 1424833_at 1424834_s_at 1427287_s_at 1427693_at 1444418_at 1444551_at 1447328_at | 18943 | -0.384 | 0.0637 | No | ||
| 51 | ATP6V1C1 | 1419544_at 1419545_a_at 1419546_at | 19313 | -0.466 | 0.0576 | No | ||
| 52 | TMEM1 | 1435430_at | 19349 | -0.473 | 0.0668 | No | ||
| 53 | NNT | 1416105_at 1432608_at 1456573_x_at | 20107 | -0.700 | 0.0483 | No | ||
| 54 | ATP6V0C | 1416392_a_at 1435732_x_at 1438925_x_at | 20187 | -0.731 | 0.0615 | No | ||
| 55 | SLC30A4 | 1418843_at | 20348 | -0.798 | 0.0726 | No |