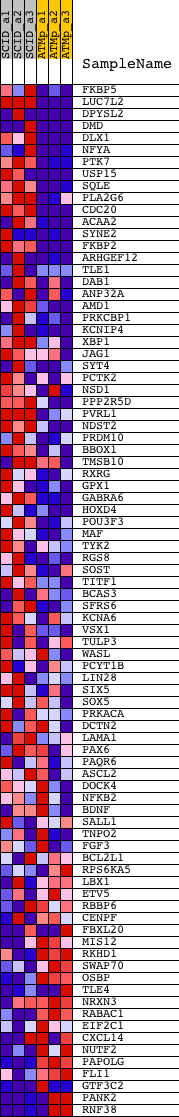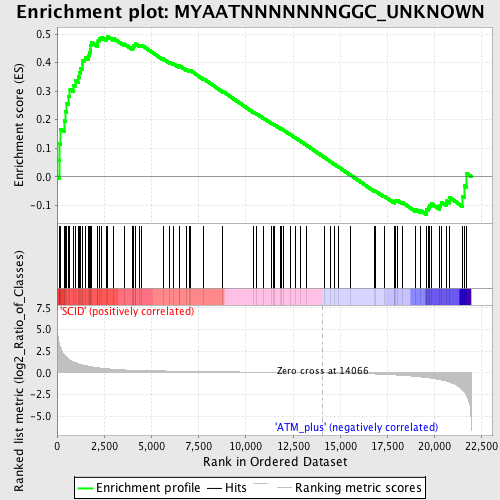
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus_repos |
| Phenotype | phenotype_SCID_versus_ATM_plus.cls#SCID_versus_ATM_plus_repos |
| Upregulated in class | SCID |
| GeneSet | MYAATNNNNNNNGGC_UNKNOWN |
| Enrichment Score (ES) | 0.49270183 |
| Normalized Enrichment Score (NES) | 1.4720736 |
| Nominal p-value | 0.013833992 |
| FDR q-value | 0.14005554 |
| FWER p-Value | 0.938 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | FKBP5 | 1416125_at 1447736_at 1448231_at 1458089_at | 110 | 3.370 | 0.0567 | Yes | ||
| 2 | LUC7L2 | 1423251_at 1427087_at 1436165_at 1436766_at 1436767_at 1437761_at 1458423_at | 124 | 3.275 | 0.1162 | Yes | ||
| 3 | DPYSL2 | 1433770_at 1450502_at 1458188_at 1458632_at | 189 | 2.899 | 0.1664 | Yes | ||
| 4 | DMD | 1417307_at 1430320_at 1446156_at 1448665_at 1457022_at | 376 | 2.102 | 0.1964 | Yes | ||
| 5 | DLX1 | 1449470_at | 454 | 1.923 | 0.2281 | Yes | ||
| 6 | NFYA | 1422082_a_at 1427808_at 1452560_a_at | 518 | 1.784 | 0.2579 | Yes | ||
| 7 | PTK7 | 1452589_at | 613 | 1.592 | 0.2828 | Yes | ||
| 8 | USP15 | 1436891_at 1446186_at 1454036_a_at 1455842_x_at | 681 | 1.471 | 0.3067 | Yes | ||
| 9 | SQLE | 1415993_at | 882 | 1.236 | 0.3202 | Yes | ||
| 10 | PLA2G6 | 1422147_a_at 1431278_s_at | 953 | 1.187 | 0.3388 | Yes | ||
| 11 | CDC20 | 1416664_at 1439377_x_at 1439394_x_at | 1125 | 1.041 | 0.3500 | Yes | ||
| 12 | ACAA2 | 1428145_at 1428146_s_at 1455061_a_at | 1176 | 1.007 | 0.3662 | Yes | ||
| 13 | SYNE2 | 1427982_s_at 1438585_at 1442285_at 1447159_at 1459322_at | 1249 | 0.965 | 0.3806 | Yes | ||
| 14 | FKBP2 | 1450694_at | 1337 | 0.919 | 0.3935 | Yes | ||
| 15 | ARHGEF12 | 1423902_s_at 1446965_at 1451159_at 1453853_a_at | 1364 | 0.903 | 0.4088 | Yes | ||
| 16 | TLE1 | 1422751_at 1434033_at | 1476 | 0.850 | 0.4193 | Yes | ||
| 17 | DAB1 | 1421100_a_at 1427307_a_at 1427308_at 1435577_at 1435578_s_at | 1662 | 0.759 | 0.4248 | Yes | ||
| 18 | ANP32A | 1421918_at 1434555_at 1450407_a_at | 1702 | 0.739 | 0.4365 | Yes | ||
| 19 | AMD1 | 1448484_at | 1771 | 0.713 | 0.4465 | Yes | ||
| 20 | PRKCBP1 | 1426614_at 1429415_at 1445827_at 1456143_at 1458617_at | 1785 | 0.710 | 0.4589 | Yes | ||
| 21 | KCNIP4 | 1437631_at 1450370_a_at 1451840_at | 1804 | 0.702 | 0.4710 | Yes | ||
| 22 | XBP1 | 1420011_s_at 1420012_at 1420886_a_at 1437223_s_at | 2129 | 0.598 | 0.4671 | Yes | ||
| 23 | JAG1 | 1421105_at 1421106_at 1434070_at | 2164 | 0.588 | 0.4763 | Yes | ||
| 24 | SYT4 | 1415844_at 1415845_at | 2221 | 0.575 | 0.4843 | Yes | ||
| 25 | PCTK2 | 1435143_at 1446130_at 1446272_at | 2335 | 0.544 | 0.4891 | Yes | ||
| 26 | NSD1 | 1420881_at 1435088_at 1440972_at 1456916_at | 2630 | 0.483 | 0.4845 | Yes | ||
| 27 | PPP2R5D | 1450560_a_at | 2644 | 0.481 | 0.4927 | Yes | ||
| 28 | PVRL1 | 1438111_at 1438421_at 1440131_at 1446488_at 1450819_at | 2967 | 0.425 | 0.4858 | No | ||
| 29 | NDST2 | 1417931_at | 3557 | 0.345 | 0.4651 | No | ||
| 30 | PRDM10 | 1441764_at 1444812_at | 3974 | 0.310 | 0.4518 | No | ||
| 31 | BBOX1 | 1419618_at | 4040 | 0.305 | 0.4544 | No | ||
| 32 | TMSB10 | 1417219_s_at 1436682_at 1436902_x_at 1437185_s_at 1455946_x_at | 4049 | 0.304 | 0.4596 | No | ||
| 33 | RXRG | 1418782_at | 4125 | 0.299 | 0.4617 | No | ||
| 34 | GPX1 | 1460671_at | 4159 | 0.297 | 0.4656 | No | ||
| 35 | GABRA6 | 1417121_at 1451706_a_at | 4384 | 0.282 | 0.4605 | No | ||
| 36 | HOXD4 | 1450209_at | 4490 | 0.276 | 0.4608 | No | ||
| 37 | POU3F3 | 1422331_at 1435197_at | 5623 | 0.226 | 0.4131 | No | ||
| 38 | MAF | 1435828_at 1437473_at 1444073_at 1447849_s_at 1447945_at 1456060_at | 5957 | 0.213 | 0.4018 | No | ||
| 39 | TYK2 | 1417306_at | 6139 | 0.207 | 0.3973 | No | ||
| 40 | RGS8 | 1453060_at | 6455 | 0.197 | 0.3865 | No | ||
| 41 | SOST | 1421245_at 1436240_at 1450179_at | 6468 | 0.196 | 0.3895 | No | ||
| 42 | TITF1 | 1422346_at | 6856 | 0.183 | 0.3752 | No | ||
| 43 | BCAS3 | 1423528_at 1428454_at 1457817_at 1458557_at | 7006 | 0.178 | 0.3716 | No | ||
| 44 | SFRS6 | 1416720_at 1416721_s_at 1447898_s_at 1448454_at | 7077 | 0.176 | 0.3717 | No | ||
| 45 | KCNA6 | 1421568_at 1441049_at 1456954_at | 7776 | 0.155 | 0.3426 | No | ||
| 46 | VSX1 | 1422104_at 1450487_at | 8784 | 0.130 | 0.2988 | No | ||
| 47 | TULP3 | 1418251_at 1449008_at | 10401 | 0.091 | 0.2266 | No | ||
| 48 | WASL | 1426776_at 1426777_a_at 1432155_at 1439832_at 1452193_a_at | 10555 | 0.087 | 0.2211 | No | ||
| 49 | PCYT1B | 1436124_at 1437648_at | 10956 | 0.077 | 0.2043 | No | ||
| 50 | LIN28 | 1437752_at | 11363 | 0.069 | 0.1869 | No | ||
| 51 | SIX5 | 1427560_at | 11466 | 0.066 | 0.1835 | No | ||
| 52 | SOX5 | 1423500_a_at 1432189_a_at 1432190_at 1440827_x_at 1446461_at 1452511_at 1459261_at | 11524 | 0.065 | 0.1821 | No | ||
| 53 | PRKACA | 1447720_x_at 1450519_a_at | 11849 | 0.056 | 0.1683 | No | ||
| 54 | DCTN2 | 1424461_at | 11901 | 0.055 | 0.1669 | No | ||
| 55 | LAMA1 | 1418153_at | 11993 | 0.052 | 0.1637 | No | ||
| 56 | PAX6 | 1419271_at 1425960_s_at 1437816_at 1452526_a_at 1456342_at | 12344 | 0.045 | 0.1485 | No | ||
| 57 | PAQR6 | 1455216_at | 12603 | 0.039 | 0.1374 | No | ||
| 58 | ASCL2 | 1422396_s_at 1432018_at 1460514_s_at | 12915 | 0.031 | 0.1238 | No | ||
| 59 | DOCK4 | 1431114_at 1436405_at 1441462_at 1441469_at 1446283_at 1457146_at 1459279_at | 13206 | 0.024 | 0.1109 | No | ||
| 60 | NFKB2 | 1425902_a_at 1429128_x_at | 14187 | -0.004 | 0.0661 | No | ||
| 61 | BDNF | 1422168_a_at 1422169_a_at | 14468 | -0.012 | 0.0536 | No | ||
| 62 | SALL1 | 1450489_at | 14688 | -0.020 | 0.0439 | No | ||
| 63 | TNPO2 | 1425592_at 1425593_at 1437283_at | 14901 | -0.026 | 0.0347 | No | ||
| 64 | FGF3 | 1422923_at 1441914_x_at | 15532 | -0.050 | 0.0068 | No | ||
| 65 | BCL2L1 | 1420887_a_at 1420888_at 1426050_at 1426191_a_at | 16797 | -0.118 | -0.0489 | No | ||
| 66 | RPS6KA5 | 1431050_at 1437625_at 1439004_at 1440343_at | 16864 | -0.123 | -0.0497 | No | ||
| 67 | LBX1 | 1422323_a_at 1427400_at | 17313 | -0.162 | -0.0672 | No | ||
| 68 | ETV5 | 1420998_at 1428142_at 1450082_s_at | 17851 | -0.217 | -0.0878 | No | ||
| 69 | RBBP6 | 1425114_at 1425115_at 1425421_at 1426487_a_at 1453611_at | 17893 | -0.223 | -0.0856 | No | ||
| 70 | CENPF | 1427161_at 1452334_at 1458447_at | 17928 | -0.227 | -0.0830 | No | ||
| 71 | FBXL20 | 1442464_at 1445575_at 1452826_s_at 1456378_s_at | 18027 | -0.243 | -0.0830 | No | ||
| 72 | MIS12 | 1424717_at | 18292 | -0.280 | -0.0900 | No | ||
| 73 | RKHD1 | 1437627_at | 18962 | -0.387 | -0.1135 | No | ||
| 74 | SWAP70 | 1423543_at 1434225_at 1440809_at | 19226 | -0.445 | -0.1174 | No | ||
| 75 | OSBP | 1460350_at | 19545 | -0.524 | -0.1223 | No | ||
| 76 | TLE4 | 1425650_at 1430384_at 1450853_at | 19550 | -0.526 | -0.1129 | No | ||
| 77 | NRXN3 | 1419825_at 1431153_at 1432931_at 1433788_at 1438193_at 1439629_at 1442423_at 1444700_at 1456137_at 1460101_at | 19658 | -0.551 | -0.1077 | No | ||
| 78 | RABAC1 | 1427773_a_at | 19734 | -0.575 | -0.1006 | No | ||
| 79 | EIF2C1 | 1434331_at | 19806 | -0.597 | -0.0929 | No | ||
| 80 | CXCL14 | 1418456_a_at 1418457_at | 20268 | -0.770 | -0.0999 | No | ||
| 81 | NUTF2 | 1434367_s_at 1447440_at 1449017_at | 20332 | -0.792 | -0.0882 | No | ||
| 82 | PAPOLG | 1427241_at 1446307_at | 20624 | -0.934 | -0.0844 | No | ||
| 83 | FLI1 | 1422024_at 1433512_at 1441584_at | 20787 | -1.055 | -0.0725 | No | ||
| 84 | GTF3C2 | 1428448_a_at 1428449_at 1440307_at 1452780_at 1452781_a_at | 21488 | -2.002 | -0.0678 | No | ||
| 85 | PANK2 | 1431948_a_at 1443846_x_at 1452733_at 1458358_at | 21576 | -2.210 | -0.0313 | No | ||
| 86 | RNF38 | 1428101_at 1431993_a_at 1445272_at | 21689 | -2.605 | 0.0113 | No |