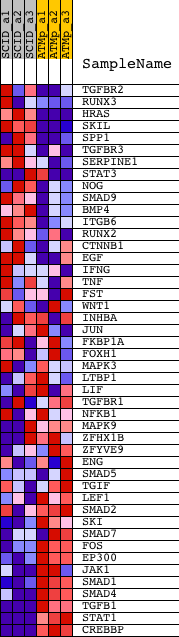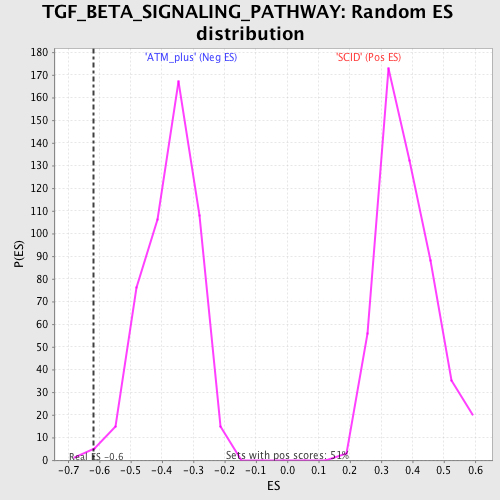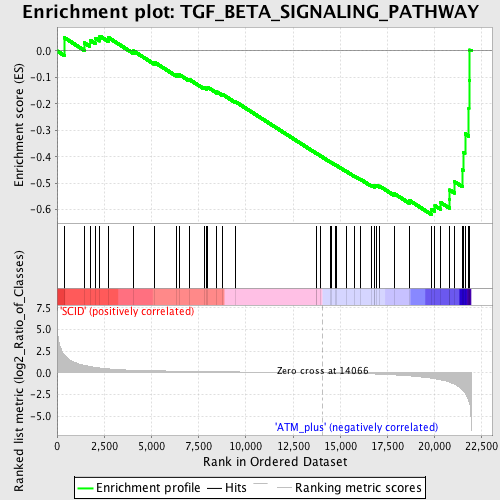
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus_repos |
| Phenotype | phenotype_SCID_versus_ATM_plus.cls#SCID_versus_ATM_plus_repos |
| Upregulated in class | ATM_plus |
| GeneSet | TGF_BETA_SIGNALING_PATHWAY |
| Enrichment Score (ES) | -0.61809057 |
| Normalized Enrichment Score (NES) | -1.6668917 |
| Nominal p-value | 0.004056795 |
| FDR q-value | 0.19791101 |
| FWER p-Value | 0.977 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | TGFBR2 | 1422019_at 1425444_a_at 1426397_at 1443115_at 1458607_at | 393 | 2.063 | 0.0498 | No | ||
| 2 | RUNX3 | 1421467_at 1440275_at | 1440 | 0.868 | 0.0306 | No | ||
| 3 | HRAS | 1422407_s_at 1424132_at | 1741 | 0.725 | 0.0407 | No | ||
| 4 | SKIL | 1422054_a_at 1452214_at | 2020 | 0.629 | 0.0487 | No | ||
| 5 | SPP1 | 1449254_at | 2264 | 0.563 | 0.0561 | No | ||
| 6 | TGFBR3 | 1425620_at 1433795_at 1440041_at 1442332_at 1447314_at 1459355_at | 2724 | 0.465 | 0.0504 | No | ||
| 7 | SERPINE1 | 1419149_at | 4044 | 0.305 | 0.0001 | No | ||
| 8 | STAT3 | 1424272_at 1426587_a_at 1459961_a_at 1460700_at | 5150 | 0.246 | -0.0422 | No | ||
| 9 | NOG | 1422300_at | 6324 | 0.201 | -0.0892 | No | ||
| 10 | SMAD9 | 1450265_at | 6460 | 0.196 | -0.0889 | No | ||
| 11 | BMP4 | 1422912_at | 6993 | 0.179 | -0.1074 | No | ||
| 12 | ITGB6 | 1422983_at 1432281_a_at | 7789 | 0.155 | -0.1386 | No | ||
| 13 | RUNX2 | 1424704_at 1425389_a_at 1426034_a_at | 7937 | 0.151 | -0.1404 | No | ||
| 14 | CTNNB1 | 1420811_a_at 1430533_a_at 1450008_a_at | 7984 | 0.149 | -0.1376 | No | ||
| 15 | EGF | 1418093_a_at | 8460 | 0.138 | -0.1547 | No | ||
| 16 | IFNG | 1425947_at | 8785 | 0.130 | -0.1653 | No | ||
| 17 | TNF | 1419607_at | 9455 | 0.113 | -0.1921 | No | ||
| 18 | FST | 1421365_at 1434458_at | 13754 | 0.008 | -0.3882 | No | ||
| 19 | WNT1 | 1425377_at | 13758 | 0.008 | -0.3881 | No | ||
| 20 | INHBA | 1422053_at 1458291_at | 13937 | 0.004 | -0.3961 | No | ||
| 21 | JUN | 1417409_at 1448694_at | 14497 | -0.014 | -0.4212 | No | ||
| 22 | FKBP1A | 1416036_at 1438795_x_at 1438958_x_at 1448184_at 1456196_x_at | 14549 | -0.015 | -0.4230 | No | ||
| 23 | FOXH1 | 1422212_at 1422213_s_at 1437779_at | 14762 | -0.022 | -0.4320 | No | ||
| 24 | MAPK3 | 1427060_at | 14783 | -0.022 | -0.4321 | No | ||
| 25 | LTBP1 | 1419786_at 1440678_at 1446232_at 1447547_at 1448870_at 1457852_at 1458739_at | 15313 | -0.041 | -0.4549 | No | ||
| 26 | LIF | 1421206_at 1421207_at 1450160_at | 15753 | -0.059 | -0.4731 | No | ||
| 27 | TGFBR1 | 1420893_a_at 1420894_at 1420895_at 1446946_at | 16054 | -0.073 | -0.4844 | No | ||
| 28 | NFKB1 | 1427705_a_at 1442949_at | 16630 | -0.107 | -0.5071 | No | ||
| 29 | MAPK9 | 1421876_at 1421877_at 1421878_at | 16802 | -0.119 | -0.5110 | No | ||
| 30 | ZFHX1B | 1422748_at 1434298_at 1442393_at 1445518_at 1454200_at 1456389_at | 16812 | -0.119 | -0.5075 | No | ||
| 31 | ZFYVE9 | 1440348_at | 16891 | -0.125 | -0.5070 | No | ||
| 32 | ENG | 1417271_a_at 1432176_a_at | 17067 | -0.140 | -0.5104 | No | ||
| 33 | SMAD5 | 1421047_at 1433641_at 1451873_a_at | 17862 | -0.219 | -0.5395 | No | ||
| 34 | TGIF | 1422286_a_at | 18672 | -0.332 | -0.5655 | No | ||
| 35 | LEF1 | 1421299_a_at 1436398_at 1440760_at | 19824 | -0.603 | -0.5983 | Yes | ||
| 36 | SMAD2 | 1420634_a_at 1458304_at | 20011 | -0.663 | -0.5850 | Yes | ||
| 37 | SKI | 1426373_at 1429192_at 1444785_at 1456547_at | 20313 | -0.785 | -0.5730 | Yes | ||
| 38 | SMAD7 | 1423389_at 1440952_at 1443771_x_at | 20786 | -1.054 | -0.5599 | Yes | ||
| 39 | FOS | 1423100_at | 20795 | -1.059 | -0.5254 | Yes | ||
| 40 | EP300 | 1434765_at | 21052 | -1.301 | -0.4944 | Yes | ||
| 41 | JAK1 | 1433803_at 1433804_at 1433805_at | 21465 | -1.953 | -0.4490 | Yes | ||
| 42 | SMAD1 | 1416081_at 1448208_at 1459843_s_at | 21506 | -2.046 | -0.3836 | Yes | ||
| 43 | SMAD4 | 1422485_at 1422486_a_at 1422487_at 1444205_at | 21604 | -2.282 | -0.3130 | Yes | ||
| 44 | TGFB1 | 1420653_at 1445360_at | 21809 | -3.227 | -0.2163 | Yes | ||
| 45 | STAT1 | 1420915_at 1440481_at 1450033_a_at 1450034_at | 21816 | -3.247 | -0.1098 | Yes | ||
| 46 | CREBBP | 1434633_at 1435224_at 1436983_at 1444856_at 1457641_at 1459804_at | 21856 | -3.508 | 0.0037 | Yes |