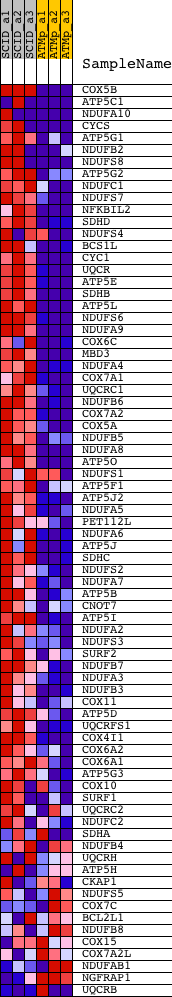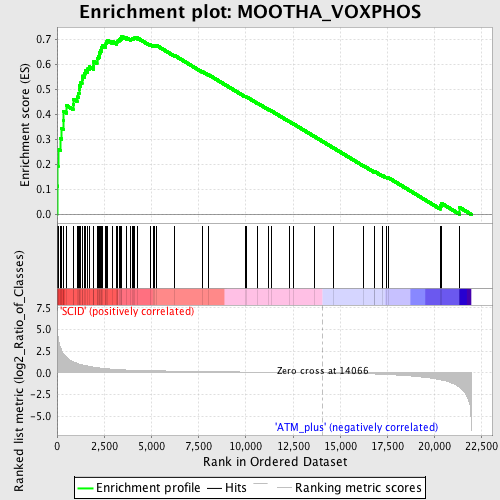
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_01_SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus.phenotype_SCID_versus_ATM_plus.cls #SCID_versus_ATM_plus_repos |
| Phenotype | phenotype_SCID_versus_ATM_plus.cls#SCID_versus_ATM_plus_repos |
| Upregulated in class | SCID |
| GeneSet | MOOTHA_VOXPHOS |
| Enrichment Score (ES) | 0.7142997 |
| Normalized Enrichment Score (NES) | 2.0800972 |
| Nominal p-value | 0.0 |
| FDR q-value | 2.5522112E-4 |
| FWER p-Value | 0.0010 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | COX5B | 1416902_a_at 1435613_x_at 1436149_at 1448514_at 1454716_x_at 1456588_x_at | 1 | 6.901 | 0.1141 | Yes | ||
| 2 | ATP5C1 | 1416058_s_at 1438809_at | 29 | 4.740 | 0.1913 | Yes | ||
| 3 | NDUFA10 | 1418068_at 1448934_at | 51 | 4.175 | 0.2594 | Yes | ||
| 4 | CYCS | 1422484_at 1445484_at | 164 | 3.037 | 0.3045 | Yes | ||
| 5 | ATP5G1 | 1416020_a_at 1444874_at | 227 | 2.644 | 0.3454 | Yes | ||
| 6 | NDUFB2 | 1416834_x_at 1442658_at 1448483_a_at 1453806_at | 328 | 2.212 | 0.3775 | Yes | ||
| 7 | NDUFS8 | 1423907_a_at 1423908_at 1434212_at 1434213_x_at 1434579_x_at 1455283_x_at | 337 | 2.196 | 0.4134 | Yes | ||
| 8 | ATP5G2 | 1415980_at 1456128_at | 502 | 1.824 | 0.4361 | Yes | ||
| 9 | NDUFC1 | 1416285_at 1448284_a_at | 847 | 1.273 | 0.4414 | Yes | ||
| 10 | NDUFS7 | 1424313_a_at 1451312_at | 864 | 1.252 | 0.4614 | Yes | ||
| 11 | NFKBIL2 | 1428661_at | 1059 | 1.093 | 0.4706 | Yes | ||
| 12 | SDHD | 1428235_at 1437489_x_at | 1135 | 1.031 | 0.4843 | Yes | ||
| 13 | NDUFS4 | 1418117_at 1438166_x_at 1444464_at 1448959_at 1459760_at | 1166 | 1.014 | 0.4997 | Yes | ||
| 14 | BCS1L | 1451541_at | 1183 | 1.005 | 0.5156 | Yes | ||
| 15 | CYC1 | 1416604_at | 1259 | 0.960 | 0.5280 | Yes | ||
| 16 | UQCR | 1448292_at | 1321 | 0.925 | 0.5405 | Yes | ||
| 17 | ATP5E | 1416567_s_at | 1345 | 0.915 | 0.5546 | Yes | ||
| 18 | SDHB | 1418005_at | 1446 | 0.863 | 0.5643 | Yes | ||
| 19 | ATP5L | 1448203_at | 1485 | 0.846 | 0.5766 | Yes | ||
| 20 | NDUFS6 | 1433603_at | 1631 | 0.777 | 0.5828 | Yes | ||
| 21 | NDUFA9 | 1416663_at | 1706 | 0.737 | 0.5916 | Yes | ||
| 22 | COX6C | 1415970_at 1434491_a_at | 1922 | 0.661 | 0.5927 | Yes | ||
| 23 | MBD3 | 1417728_at | 1940 | 0.655 | 0.6028 | Yes | ||
| 24 | NDUFA4 | 1424085_at | 1943 | 0.654 | 0.6135 | Yes | ||
| 25 | COX7A1 | 1418709_at | 2114 | 0.603 | 0.6157 | Yes | ||
| 26 | UQCRC1 | 1428782_a_at | 2115 | 0.601 | 0.6256 | Yes | ||
| 27 | NDUFB6 | 1434056_a_at 1434057_at | 2181 | 0.583 | 0.6323 | Yes | ||
| 28 | COX7A2 | 1416970_a_at 1416971_at | 2232 | 0.572 | 0.6395 | Yes | ||
| 29 | COX5A | 1415933_a_at 1439267_x_at 1448153_at | 2259 | 0.565 | 0.6477 | Yes | ||
| 30 | NDUFB5 | 1417102_a_at 1448589_at | 2285 | 0.558 | 0.6557 | Yes | ||
| 31 | NDUFA8 | 1423692_at | 2355 | 0.539 | 0.6615 | Yes | ||
| 32 | ATP5O | 1416278_a_at | 2361 | 0.538 | 0.6702 | Yes | ||
| 33 | NDUFS1 | 1425143_a_at 1453354_at | 2398 | 0.531 | 0.6773 | Yes | ||
| 34 | ATP5F1 | 1426742_at 1433562_s_at | 2558 | 0.497 | 0.6783 | Yes | ||
| 35 | ATP5J2 | 1416269_at 1435395_s_at | 2586 | 0.491 | 0.6852 | Yes | ||
| 36 | NDUFA5 | 1417285_a_at 1417286_at | 2632 | 0.483 | 0.6911 | Yes | ||
| 37 | PET112L | 1424282_at 1444502_at | 2683 | 0.471 | 0.6966 | Yes | ||
| 38 | NDUFA6 | 1448427_at | 2922 | 0.431 | 0.6928 | Yes | ||
| 39 | ATP5J | 1416143_at | 3166 | 0.393 | 0.6882 | Yes | ||
| 40 | SDHC | 1435986_x_at 1448630_a_at | 3176 | 0.391 | 0.6943 | Yes | ||
| 41 | NDUFS2 | 1451096_at | 3223 | 0.385 | 0.6986 | Yes | ||
| 42 | NDUFA7 | 1422976_x_at 1428360_x_at 1436567_a_at 1450818_a_at 1455749_x_at | 3311 | 0.374 | 0.7008 | Yes | ||
| 43 | ATP5B | 1416829_at | 3364 | 0.368 | 0.7045 | Yes | ||
| 44 | CNOT7 | 1423641_s_at 1430519_a_at 1460665_a_at | 3412 | 0.362 | 0.7083 | Yes | ||
| 45 | ATP5I | 1422525_at 1434053_x_at 1450640_x_at | 3413 | 0.362 | 0.7143 | Yes | ||
| 46 | NDUFA2 | 1417368_s_at | 3685 | 0.334 | 0.7074 | No | ||
| 47 | NDUFS3 | 1423737_at | 3907 | 0.316 | 0.7026 | No | ||
| 48 | SURF2 | 1433609_s_at 1453117_at | 3967 | 0.311 | 0.7050 | No | ||
| 49 | NDUFB7 | 1416417_a_at 1448331_at | 4053 | 0.304 | 0.7061 | No | ||
| 50 | NDUFA3 | 1428464_at 1430149_at 1452790_x_at | 4119 | 0.300 | 0.7081 | No | ||
| 51 | NDUFB3 | 1416547_at | 4243 | 0.292 | 0.7073 | No | ||
| 52 | COX11 | 1429188_at 1457846_at | 4930 | 0.256 | 0.6802 | No | ||
| 53 | ATP5D | 1423716_s_at 1456580_s_at | 5080 | 0.249 | 0.6775 | No | ||
| 54 | UQCRFS1 | 1450968_at | 5182 | 0.245 | 0.6769 | No | ||
| 55 | COX4I1 | 1448322_a_at | 5260 | 0.241 | 0.6774 | No | ||
| 56 | COX6A2 | 1417607_at 1457633_x_at | 6232 | 0.203 | 0.6363 | No | ||
| 57 | COX6A1 | 1417417_a_at | 7695 | 0.157 | 0.5721 | No | ||
| 58 | ATP5G3 | 1454661_at | 8034 | 0.148 | 0.5591 | No | ||
| 59 | COX10 | 1429329_at 1446223_at 1459977_x_at | 9962 | 0.101 | 0.4726 | No | ||
| 60 | SURF1 | 1450561_a_at | 10054 | 0.098 | 0.4700 | No | ||
| 61 | UQCRC2 | 1428631_a_at 1435757_a_at | 10607 | 0.086 | 0.4462 | No | ||
| 62 | NDUFC2 | 1431670_at | 11217 | 0.072 | 0.4195 | No | ||
| 63 | SDHA | 1426688_at 1426689_s_at 1445317_at | 11345 | 0.069 | 0.4149 | No | ||
| 64 | NDUFB4 | 1428075_at 1428076_s_at 1432427_at | 12291 | 0.046 | 0.3724 | No | ||
| 65 | UQCRH | 1452133_at 1453229_s_at | 12494 | 0.041 | 0.3639 | No | ||
| 66 | ATP5H | 1423676_at 1435112_a_at | 13608 | 0.012 | 0.3131 | No | ||
| 67 | CKAP1 | 1448533_at | 14622 | -0.017 | 0.2671 | No | ||
| 68 | NDUFS5 | 1416494_at 1416495_s_at | 16242 | -0.082 | 0.1944 | No | ||
| 69 | COX7C | 1448112_at 1459884_at 1459885_s_at | 16794 | -0.118 | 0.1711 | No | ||
| 70 | BCL2L1 | 1420887_a_at 1420888_at 1426050_at 1426191_a_at | 16797 | -0.118 | 0.1730 | No | ||
| 71 | NDUFB8 | 1448198_a_at 1458404_at | 17221 | -0.154 | 0.1562 | No | ||
| 72 | COX15 | 1426693_x_at 1437982_x_at 1452146_a_at 1460376_a_at | 17422 | -0.173 | 0.1499 | No | ||
| 73 | COX7A2L | 1421772_a_at 1432263_a_at 1432264_x_at | 17553 | -0.185 | 0.1470 | No | ||
| 74 | NDUFAB1 | 1428159_s_at 1428160_at 1435934_at 1447919_x_at 1453565_at 1458678_at | 20293 | -0.778 | 0.0346 | No | ||
| 75 | NGFRAP1 | 1428842_a_at | 20339 | -0.794 | 0.0457 | No | ||
| 76 | UQCRB | 1416337_at 1455997_a_at | 21322 | -1.651 | 0.0281 | No |